Abstract
Purpose
Breast cancer is a molecularly heterogeneous disease, and multiple genetic variants contribute to its development and prognosis. Most of previous genome-wide association studies (GWASs) and polygenic risk scores (PRSs) analyses focused on studying breast cancers of Caucasian populations, which may not be applicable to other population. Therefore, we conducted the largest breast cancer cohort of Taiwanese population to fill in the knowledge gap.
Methods
A total of 152,534 Participants recruited by China Medical University Hospital between 2003 and 2019 were filtered by several patient selection criteria and GWAS quality control steps, resulting in the inclusion of 2496 cases and 9984 controls for this study. We then conducted GWAS for all breast cancers and PRS analyses for all breast cancers and the four breast cancer subtypes, including luminal A, luminal B, basal-like, and HER2-enriched.
Results
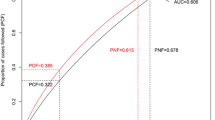
The GWAS analyses identified 113 SNPs, 50 of which were novel. The PRS models for all breast cancers and the luminal A subtype showed positively correlated trends between the PRS and the risk of developing breast cancer. The odds ratios (95% confidence intervals) for the groups with the highest PRS in all breast cancers and the luminal A subtype were 5.33 (3.79–7.66) and 3.55 (2.13–6.14), respectively.
Conclusion
In summary, we explored the association of genetic variants with breast cancer in the largest Taiwanese cohort and developed two PRS models that can predict the risk of developing any breast cancer and the luminal A subtype in Taiwanese women.







Similar content being viewed by others
Data availability
The dataset supporting the conclusions of this article is available in the China Medical Hospital repository. Anyone who is interested in accessing the data must contact China Medical Hospital thorough the corresponding author Dr. Tsai.
Code availability
Codes for data preprocessing and analysis in this study are available online at https://github.com/ychsu2014/Taiwan_2496_breast_cancers for review.
References
Wilkinson L, Gathani T (2022) Understanding breast cancer as a global health concern. Br J Radiol 95(1130):20211033. https://doi.org/10.1259/bjr.20211033
Arnold M et al (2022) Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast 66:15–23. https://doi.org/10.1016/j.breast.2022.08.010
2020 Cause of Death Statistics (2020) Available from: https://www.mohw.gov.tw/cp-5256-63399-2.html
Taiwan Cancer Registry. 2017 Available from: https://www.hpa.gov.tw/EngPages/Detail.aspx?nodeid=1061&pid=6069
Maxwell KN, Nathanson KL (2013) Common breast cancer risk variants in the post-COGS era: a comprehensive review. Breast Cancer Res 15(6):212. https://doi.org/10.1186/bcr3591
Liang B et al (2020) GWAS in cancer: progress and challenges. Mol Genet Genomics 295(3):537–561. https://doi.org/10.1007/s00438-020-01647-z
Easton DF et al (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447(7148):1087–1093. https://doi.org/10.1038/nature05887
Choi SW, Mak TS, O’Reilly PF (2020) Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 15(9):2759–2772. https://doi.org/10.1038/s41596-020-0353-1
Mars N et al (2020) The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat Commun 11(1):6383. https://doi.org/10.1038/s41467-020-19966-5
Mavaddat N et al (2015) Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. https://doi.org/10.1093/jnci/djv036
Shieh Y et al (2016) Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res Treat 159(3):513–525. https://doi.org/10.1007/s10549-016-3953-2
Kramer I et al (2020) Breast cancer polygenic risk score and contralateral breast cancer risk. Am J Hum Genet 107(5):837–848. https://doi.org/10.1016/j.ajhg.2020.09.001
Tsai CW et al (2019) Longitudinal progression trajectory of random urine creatinine as a novel predictor of ESRD among patients with CKD. Clin Chim Acta 489:144–153. https://doi.org/10.1016/j.cca.2018.12.002
Chiang HY et al (2021) Association between preoperative blood glucose level and hospital length of stay for patients undergoing appendectomy or laparoscopic cholecystectomy. Diabetes Care 44(1):107–115. https://doi.org/10.2337/dc19-0963
Chiang HY et al (2021) Electronic medical record-based deep data cleaning and phenotyping improve the diagnostic validity and mortality assessment of infective endocarditis: medical big data initiative of CMUH. Biomedicine (Taipei) 11(3):59–67. https://doi.org/10.37796/2211-8039.1267
Liang HY et al (2020) Validation and comparison of the 2003 and 2016 diastolic functional assessments for cardiovascular mortality in a large single-center cohort. J Am Soc Echocardiogr 33(4):469–480. https://doi.org/10.1016/j.echo.2019.11.013
Huang CC et al (2019) Development of a prediction model for breast cancer based on the national cancer registry in Taiwan. Breast Cancer Res 21(1):92. https://doi.org/10.1186/s13058-019-1172-6
Wei CY et al (2021) Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom Med 6(1):10. https://doi.org/10.1038/s41525-021-00178-9
Chattopadhyay A et al (2023) Multi-ethnic imputation system (MI-System): a genotype imputation server for high-dimensional data. J Biomed Inform. https://doi.org/10.1016/j.jbi.2023.104423
Chang CC et al (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. https://doi.org/10.1186/s13742-015-0047-8
Delaneau O, Zagury JF, Marchini J (2013) Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 10(1):5–6. https://doi.org/10.1038/nmeth.2307
Howie B, Marchini J, Stephens M (2011) Genotype imputation with thousands of genomes. G3 (Bethesda) 1(6):457–470. https://doi.org/10.1534/g3.111.001198
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5(6):e1000529. https://doi.org/10.1371/journal.pgen.1000529
Genomes Project C et al (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Hong EP, Park JW (2012) Sample size and statistical power calculation in genetic association studies. Genomics Inform 10(2):117–122. https://doi.org/10.5808/GI.2012.10.2.117
Baek S et al (2015) Propensity score matching: a conceptual review for radiology researchers. Korean J Radiol 16(2):286–296. https://doi.org/10.3348/kjr.2015.16.2.286
Chen JW et al (2022) Best practice guidelines for propensity score methods in medical research: consideration on theory, implementation, and reporting. A Rev Arthroscopy 38(2):632–642. https://doi.org/10.1016/j.arthro.2021.06.037
Zhao H et al (2018) A practical approach to adjusting for population stratification in genome-wide association studies: principal components and propensity scores (PCAPS). Stat Appl Genet Mol Biol. https://doi.org/10.1515/sagmb-2017-0054
Price AL et al (2010) New approaches to population stratification in genome-wide association studies. Nat Rev Genet 11(7):459–463. https://doi.org/10.1038/nrg2813
Feng Q et al (2009) A method to correct for population structure using a segregation model. BMC Proc 3(Suppl 7):104. https://doi.org/10.1186/1753-6561-3-s7-s104
Kang SJ et al (2009) Assessing the impact of global versus local ancestry in association studies. BMC Proc 3(Suppl 7):S107. https://doi.org/10.1186/1753-6561-3-s7-s107
Sherry ST et al (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29(1):308–311. https://doi.org/10.1093/nar/29.1.308
Perou CM et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752. https://doi.org/10.1038/35021093
Sorlie T et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100(14):8418–8423. https://doi.org/10.1073/pnas.0932692100
Goldhirsch A et al (2011) Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol 22(8):1736–1747. https://doi.org/10.1093/annonc/mdr304
Choi SW, O’Reilly PF (2019) PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. https://doi.org/10.1093/gigascience/giz082
Jiang P et al (2017) The protein encoded by the CCDC170 breast cancer gene functions to organize the golgi-microtubule network. EBioMedicine 22:28–43. https://doi.org/10.1016/j.ebiom.2017.06.024
Han CC et al (2016) TOX3 protein expression is correlated with pathological characteristics in breast cancer. Oncol Lett 11(3):1762–1768. https://doi.org/10.3892/ol.2016.4117
Zuo X et al (2020) The association of CASC16 variants with breast Cancer risk in a northwest Chinese female population. Mol Med 26(1):11. https://doi.org/10.1186/s10020-020-0137-7
Hu Z et al (2013) GEP100 regulates epidermal growth factor-induced MDA-MB-231 breast cancer cell invasion through the activation of Arf6/ERK/uPAR signaling pathway. Exp Cell Res 319(13):1932–1941. https://doi.org/10.1016/j.yexcr.2013.05.028
Zhou D et al (2018) Erythropoietin-producing hepatocellular A6 overexpression is a novel biomarker of poor prognosis in patients with breast cancer. Oncol Lett 15(4):5257–5263. https://doi.org/10.3892/ol.2018.7919
Li L et al (2020) Therapeutic role of recurrent ESR1-CCDC170 gene fusions in breast cancer endocrine resistance. Breast Cancer Res 22(1):84. https://doi.org/10.1186/s13058-020-01325-3
Thomassen M, Tan Q, Kruse TA (2009) Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat 113(2):239–249. https://doi.org/10.1007/s10549-008-9927-2
Lehner A et al (2013) Downregulation of serine protease HTRA1 is associated with poor survival in breast cancer. PLoS ONE 8(4):e60359. https://doi.org/10.1371/journal.pone.0060359
Pongor L et al (2015) A genome-wide approach to link genotype to clinical outcome by utilizing next generation sequencing and gene chip data of 6697 breast cancer patients. Genome Med 7:104. https://doi.org/10.1186/s13073-015-0228-1
Hsieh YC et al (2017) A polygenic risk score for breast cancer risk in a Taiwanese population. Breast Cancer Res Treat 163(1):131–138. https://doi.org/10.1007/s10549-017-4144-5
Acknowledgements
We appreciate the iHi Clinical Research and iHi Genomics Platform from the Big Data Center of China Medical University Hospital for the data exploration, administrative, and statistical analytic support.
Funding
This work was partly supported by National Science and Technology Council, Taiwan (MOST-106-2314-B-002-134-MY2, MOST-108-2314-B-002-103-MY2, and MOST-109-2314-B-002-151-MY3) and Population Health and Welfare Research Center from Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (grant number NTU-112L9004).
Author information
Authors and Affiliations
Contributions
YH: data analysis, data visualization, manuscript writing, manuscript revision; HC: data curation, data analysis, manuscript revision; CC: data curation, data analysis, manuscript revision; AC: data analysis, manuscript revision; PC: data curation; CL: data curation; HC: data curation; TYL: data curation; CK: data curation, data analysis, project design, project supervision, administrative support, manuscript revision; EYC: project supervision, administrative support; TPL: data analysis, manuscript writing, manuscript revision, project design, project supervision, administrative support; FT: data curation, project supervision, administrative support. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The study was approved by the Big Data Center of CMUH and the Research Ethical Committee/Institutional Review Board (REC/IRB) of China Medical University Hospital (CMUH105-REC3-068, CMUH107-REC3-058, CMUH110-REC1-100, and CMUH110-REC2-145). All methods were performed in accordance with the relevant guidelines and regulations of REC/IRB.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
The authors affirm that human research participants provided informed consent for publication of all the figures and tables in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hsu, YC., Chen, HL., Cheng, CF. et al. The largest genome-wide association study for breast cancer in Taiwanese Han population. Breast Cancer Res Treat 203, 291–306 (2024). https://doi.org/10.1007/s10549-023-07133-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-07133-5