Abstract
Background
Cyclin D1 overexpression may contribute to development of various cancers, including breast cancer, and thus may serve as a key cancer diagnostic marker and therapeutic target. In our previous study, we generated a cyclin D1-specific single-chain variable fragment antibody (ADκ) from a human semi-synthetic single-chain variable fragment library. ADκ specifically interacted with recombinant and endogenous cyclin D1 proteins through an unknown molecular basis to inhibit HepG2 cell growth and proliferation.
Results
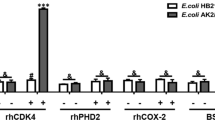
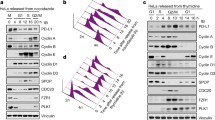
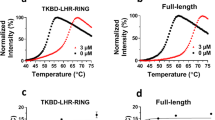
Here, using phage display and in silico protein structure modeling methods combined with cyclin D1 mutational analysis, key residues that bind to ADκ were identified. Notably, residue K112 within the cyclin box was required for cyclin D1–ADκ binding. In order to elucidate the molecular mechanism underlying ADκ anti-tumor effects, a cyclin D1-specific nuclear localization signal-containing intrabody (NLS-ADκ) was constructed. When expressed within cells, NLS-ADκ interacted specifically with cyclin D1 to significantly inhibit cell proliferation, induce G1-phase arrest, and trigger apoptosis of MCF-7 and MDA-MB-231 breast cancer cells. Moreover, the NLS–ADκ–cyclin D1 interaction blocked binding of cyclin D1 to CDK4 and inhibited RB protein phosphorylation, resulting in altered expression of downstream cell proliferation-related target genes.
Conclusion
We identified amino acid residues in cyclin D1 that may play key roles in the ADκ–cyclin D1 interaction. A nuclear localization antibody against cyclin D1 (NLS-ADκ) was constructed and successfully expressed in breast cancer cells. NLS-ADκ exerted tumor suppressor effects via blocking the binding of CDK4 to cyclin D1 and inhibiting phosphorylation of RB. The results presented here demonstrate anti-tumor potential of intrabody-based cyclin D1-targeted breast cancer therapy.







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- scFv:
-
Single-chain variable fragment
- NLS:
-
Nuclear localization signal
- E. coli :
-
Escherichia coli
- RB:
-
Retinoblastoma tumor suppressor protein
- CDK4:
-
Cyclin-dependent kinase 4
References
Matthews HK, Bertoli C, de Bruin RAM (2022) Cell cycle control in cancer. Nat Rev Mol Cell Biol 23(1):74–88. https://doi.org/10.1038/s41580-021-00404-3
Goel S, Bergholz J, Zhao J (2022) Targeting CDK4 and CDK6 in cancer. Nat Rev Cancer. https://doi.org/10.1038/s41568-022-00456-3
Fassl A, Geng Y, Sicinski P (2022) CDK4 and CDK6 kinases: from basic science to cancer therapy. Science 375(6577):eabc1495. https://doi.org/10.1126/science.abc1495
Hume S, Dianov G, Ramadan K (2020) A unified model for the G1/S cell cycle transition. Nucleic Acids Res 48(22):12483–12501. https://doi.org/10.1093/nar/gkaa1002
Qie S, Diehl JA (2020) Cyclin D degradation by E3 ligases in cancer progression and treatment. Semin Cancer Biol 67(Pt 2):159–170. https://doi.org/10.1016/j.semcancer.2020.01.012
Cemeli T et al (2019) Cytoplasmic cyclin D1 regulates glioblastoma dissemination. J Pathol 248(4):501–513. https://doi.org/10.1002/path.5277
Choi YJ et al (2012) The requirement for cyclin D function in tumor maintenance. Cancer Cell 22(4):438–451. https://doi.org/10.1016/j.ccr.2012.09.015
Deshpande A, Sicinski P, Hinds PW (2005) Cyclins and cdks in development and cancer: a perspective. Oncogene 24(17):2909–2915. https://doi.org/10.1038/sj.onc.1208618
Musgrove EA et al (2011) Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 11(8):558–572. https://doi.org/10.1038/nrc3090
Beroukhim R et al (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463(7283):899–905. https://doi.org/10.1038/nature08822
Tchakarska G, Sola B (2020) The double dealing of cyclin D1. Cell Cycle 19(2):163–178. https://doi.org/10.1080/15384101.2019.1706903
Suski JM et al (2021) Targeting cell-cycle machinery in cancer. Cancer Cell 39(6):759–778. https://doi.org/10.1016/j.ccell.2021.03.010
Marschall AL, Dübel S, Böldicke T (2015) Specific in vivo knockdown of protein function by intrabodies. MAbs 7(6):1010–35. https://doi.org/10.1080/19420862.2015.1076601
Zhang J, Yi J, Zhou P (2020) Development of bispecific antibodies in China: overview and prospects. Antib Ther 3(2):126–145. https://doi.org/10.1093/abt/tbaa011
Lin Y et al (2020) Recent progress in antitumor functions of the intracellular antibodies. Drug Discov Today 25(6):1109–1120. https://doi.org/10.1016/j.drudis.2020.02.009
Cardinale A et al (2015) Intrabody-mediated diverting of HP1β to the cytoplasm induces co-aggregation of H3–H4 histones and lamin-B receptor. Exp Cell Res 338(1):70–81. https://doi.org/10.1016/j.yexcr.2015.09.006
Cattaneo A, Chirichella M (2019) Targeting the post-translational proteome with intrabodies. Trends Biotechnol 37(6):578–591. https://doi.org/10.1016/j.tibtech.2018.11.009
Goodwin MS et al (2021) Anti-tau scFvs targeted to the cytoplasm or secretory pathway variably modify pathology and neurodegenerative phenotypes. Mol Ther 29(2):859–872. https://doi.org/10.1016/j.ymthe.2020.10.007
Zhao L et al (2019) Intrabody against prolyl hydroxylase 2 promotes angiogenesis by stabilizing hypoxia-inducible factor-1α. Sci Rep 9(1):11861. https://doi.org/10.1038/s41598-019-47891-1
Zhao L et al (2020) Intrabody against prolyl hydroxylase 2 ameliorates acetaminophen-induced acute liver injury in mice via concomitant promotion of angiogenesis and redox homeostasis. Biomed Pharmacother 123:109783. https://doi.org/10.1016/j.biopha.2019.109783
Flego M et al (2019) Intracellular human antibody fragments recognizing the VP35 protein of Zaire ebola filovirus inhibit the protein activity. BMC Biotechnol 19(1):64. https://doi.org/10.1186/s12896-019-0554-2
Wu Y et al (2020) A cyclin D1-specific single-chain variable fragment antibody that inhibits hepg2 cell growth and proliferation. Biotechnol J 15(8):e1900430. https://doi.org/10.1002/biot.201900430
Sun H et al (2021) Research advances in hydrogen-deuterium exchange mass spectrometry for protein epitope mapping. Anal Bioanal Chem 413(9):2345–2359. https://doi.org/10.1007/s00216-020-03091-9
Akbar R et al (2021) A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Rep 34(11):108856. https://doi.org/10.1016/j.celrep.2021.108856
Sivaccumar JP et al (2021) Development of a new highly selective monoclonal antibody against preferentially expressed antigen in melanoma (PRAME) and identification of the target epitope by bio-layer interferometry. Int J Mol Sci. https://doi.org/10.3390/ijms22063166
Liu Y et al (2021) Epitopes prediction for microcystin-LR by molecular docking. Ecotoxicol Environ Saf 227:112925. https://doi.org/10.1016/j.ecoenv.2021.112925
Badaya A, Sasidhar YU (2022) The role of temperature in the binding of the disordered epitope region of human thrombopoietin to antibody: a molecular dynamics simulations study. J Mol Graph Model 111:108098. https://doi.org/10.1016/j.jmgm.2021.108098
Lee VS et al (2010) Pairwise decomposition of residue interaction energies of single chain Fv with HIV-1 p17 epitope variants. Mol Immunol 47(5):982–990. https://doi.org/10.1016/j.molimm.2009.11.021
Malik A et al (2010) Modeling the three-dimensional structures of an unbound single-chain variable fragment (scFv) and its hypothetical complex with a Corynespora cassiicola toxin, cassiicolin. J Mol Model 16(12):1883–1893. https://doi.org/10.1007/s00894-010-0680-1
Johnson JL et al (2015) Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization. Acta Crystallogr D Biol Crystallogr 71(Pt 4):896–906. https://doi.org/10.1107/s1399004715001856
Day PJ et al (2009) Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci USA 106(11):4166–4170. https://doi.org/10.1073/pnas.0809645106
Yoshida A et al (2022) Non-phosphorylatable cyclin D1 mutant potentiates endometrial hyperplasia and drives carcinoma with Pten loss. Oncogene 41(15):2187–2195. https://doi.org/10.1038/s41388-022-02243-8
Fereshteh MP et al (2008) The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res 68(10):3697–3706. https://doi.org/10.1158/0008-5472.Can-07-6702
Robles AI et al (1998) Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev 12(16):2469–2474. https://doi.org/10.1101/gad.12.16.2469
Harikumar KB et al (2010) A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther 9(5):1136–1146. https://doi.org/10.1158/1535-7163.Mct-09-1145
Rasouli S, Zarghami N (2018) Synergistic growth inhibitory effects of chrysin and metformin combination on breast cancer cells through hTERT and Cyclin D1 suppression. Asian Pac J Cancer Prev 19(4):977–982. https://doi.org/10.22034/apjcp.2018.19.4.977
Sukamporn P et al (2016) Damnacanthal and its nanoformulation exhibit anti-cancer activity via cyclin D1 down-regulation. Life Sci 152:60–66. https://doi.org/10.1016/j.lfs.2016.03.038
Ashraf Y et al (2019) Immunotherapy of triple-negative breast cancer with cathepsin D-targeting antibodies. J Immunother Cancer 7(1):29. https://doi.org/10.1186/s40425-019-0498-z
Trenevska I, Li D, Banham A (2017) Therapeutic antibodies against intracellular tumor antigens. Front Immunol 8:1001. https://doi.org/10.3389/fimmu.2017.01001
Biocca S et al (1995) Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Biotechnology (NY) 13(10):1110–1115. https://doi.org/10.1038/nbt1095-1110
Wang Y, Wang L (2017) Vaccination with phage-displayed antigenic epitope. Methods Mol Biol 1625:225–235. https://doi.org/10.1007/978-1-4939-7104-6_15
Hess KL, Jewell CM (2020) Phage display as a tool for vaccine and immunotherapy development. Bioeng Transl Med 5(1):e10142. https://doi.org/10.1002/btm2.10142
Tan Y et al (2016) Advance in phage display technology for bioanalysis. Biotechnol J 11(6):732–745. https://doi.org/10.1002/biot.201500458
Cariccio VL et al (2016) Phage display revisited: epitope mapping of a monoclonal antibody directed against Neisseria meningitidis adhesin A using the PROFILER technology. MAbs 8(4):741–50. https://doi.org/10.1080/19420862.2016.1158371
Fühner V et al (2019) Epitope mapping via phage display from single-gene libraries. Methods Mol Biol 1904:353–375. https://doi.org/10.1007/978-1-4939-8958-4_17
Moreira G, Fühner V, Hust M (2018) Epitope mapping by phage display. Methods Mol Biol 1701:497–518. https://doi.org/10.1007/978-1-4939-7447-4_28
O’Donovan B et al (2020) High-resolution epitope mapping of anti-Hu and anti-Yo autoimmunity by programmable phage display. Brain Commun 2(2):fcaa059. https://doi.org/10.1093/braincomms/fcaa059
Omidfar K, Daneshpour M (2015) Advances in phage display technology for drug discovery. Expert Opin Drug Discov 10(6):651–69. https://doi.org/10.1517/17460441.2015.1037738
Ting JP et al (2018) Utilization of peptide phage display to investigate hotspots on IL-17A and what it means for drug discovery. PLoS One 13(1):e0190850. https://doi.org/10.1371/journal.pone.0190850
Du Z, Tong X, Ye X (2013) Cyclin D1 promotes cell cycle progression through enhancing NDR1/2 kinase activity independent of cyclin-dependent kinase 4. J Biol Chem 288(37):26678–26687. https://doi.org/10.1074/jbc.M113.466433
Acknowledgements
We thank Dr. Ye Sun of the School of Life Sciences, Jilin University, Changchun, China, for his assistance and suggestion with the molecular model.
Funding
This work was supported by the Natural Science Foundation of Jilin Province, China (Grant No. 20210101003JC) and the National Natural Science Foundation of China (Grant Nos. 31570934, 31170882, 81903102).
Author information
Authors and Affiliations
Contributions
JZ and YW designed and performed all experiments and various analyses. TX, CC, and TZ performed cell experiments, data collection, and analysis. ZG and SH constructed and extracted plasmids and performed data collection and analysis. ZR and XY performed protein preparation. JZ, YW, FY, and GL designed the experiments, interpreted the data, and wrote the manuscript. GL contributed to the study conception and design, supervised the study, and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Wu, Y., Xiao, T. et al. A specific anti-cyclin D1 intrabody represses breast cancer cell proliferation by interrupting the cyclin D1–CDK4 interaction. Breast Cancer Res Treat 198, 555–568 (2023). https://doi.org/10.1007/s10549-023-06866-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-06866-7