Abstract
Purpose
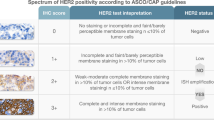
Currently, the most commonly applied method for the determination of breast cancer subtypes is to test estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki67 by immunohistochemistry (IHC). However, the IHC method has substantial intraobserver and interobserver variability. ESR1, PGR, ERBB2, and MKi67 mRNA tests by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay may improve the diagnostic objectivity and efficiency. Here, we compared the concordance between RT-qPCR and IHC for assessment of the same biomarkers and evaluated the subtypes.
Methods
A total of 265 eligible cases were divided into a training cohort and a validation cohort, and the expressions of ER/ESR1, PR/PGR, HER2/ERBB2, and Ki67/MKI67 were tested by IHC and RT-qPCR. Then, the appropriate cutoff of RT-qPCR was calculated in the training cohort. The concordance between RT-qPCR and IHC was calculated for individual marker. In addition, we investigated the subtypes based on the RT-qPCR results.
Results
The Spearman correlation coefficients between ER/ESR1, PR/PGR, HER2/ERBB2, and Ki67/MKI67 by IHC and RT-qPCR were 0.768, 0.699, 0.762, and 0.387, respectively. The cutoff values for the RT-qPCR assay of ESR1 (1%), PGR (1%), ERBB2, and MKi67 (14%) were 35.539, 32.139, 36.398, and 29.176, respectively. The overall percent agreement (OPA) between ER/ESR1, PR/PGR, HER2/ERBB2, and Ki67/MKI67 by IHC and RT-qPCR was 92.48%, 73.68%, 92.80%, and 74.44%, respectively. A total of 224 (84.53%) specimens were concordant for the breast cancer subtypes (IHC-based type) by RT-qPCR.
Conclusion
Evaluation of breast cancer biomarker status by RT-qPCR was highly concordant with IHC. RT-qPCR can be used as a supplementary method to detect molecular markers of breast cancer.




Similar content being viewed by others
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ESR1/ER:
-
Estrogen receptor alpha
- PGR/PR:
-
Progesterone receptor
- ERBB2/HER2:
-
Human epidermal growth factor receptor 2
- MKI67/Ki67:
-
Marker of proliferation Ki67
- mRNA:
-
Messenger ribonucleic acid
- IHC:
-
Immunohistochemistry
- FISH:
-
Fluorescence in situ hybridization
- RT-qPCR:
-
Reverse transcription-quantitative polymerase chain reaction
- FFPE:
-
Formalin-fixed paraffin-embedded
- GOI:
-
Genes of interest
- Cq:
-
Quantification cycle
- REF:
-
Reference genes
- Pc:
-
Positive control
- ROC:
-
Receiver operating characteristics
- OPA:
-
Overall percent agreement
- PPA:
-
Positive percent agreement
- NPA:
-
Negative percent agreement
- PPV:
-
Positive predictive value
- NPV:
-
Negative predictive value
- CI:
-
Confidence intervals
- FN:
-
False negative
- FP:
-
False positive
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71(1):7–33. https://doi.org/10.3322/caac.21654
Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H et al (2016) Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 7:11579. https://doi.org/10.1038/ncomms11579
Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD et al (2020) The single-cell pathology landscape of breast cancer. Nature 578(7796):615–620. https://doi.org/10.1038/s41586-019-1876-x
Liang Y, Zhang H, Song X, Yang Q (2020) Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol 60:14–27. https://doi.org/10.1016/j.semcancer.2019.08.012
Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M et al (2015) Tailoring therapies–improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol 26(8):1533–1546. https://doi.org/10.1093/annonc/mdv221
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752. https://doi.org/10.1038/35021093
Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL et al (2020) Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J Clin Oncol 38(12):1346–1366. https://doi.org/10.1200/jco.19.02309
Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS et al (2018) Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical practice guideline focused update. J Clin Oncol 36(20):2105–2122. https://doi.org/10.1200/jco.2018.77.8738
Nielsen TO, Leung SCY, Rimm DL, Dodson A, Acs B, Badve S et al (2021) Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in Breast Cancer Working Group. J Natl Cancer Inst 113(7):808–819. https://doi.org/10.1093/jnci/djaa201
Lee M, Lee CS, Tan PH (2013) Hormone receptor expression in breast cancer: postanalytical issues. J Clin Pathol 66(6):478–484. https://doi.org/10.1136/jclinpath-2012-201148
Orlando L, Viale G, Bria E, Lutrino ES, Sperduti I, Carbognin L et al (2016) Discordance in pathology report after central pathology review: Implications for breast cancer adjuvant treatment. Breast 30:151–155. https://doi.org/10.1016/j.breast.2016.09.015
Reisenbichler ES, Lester SC, Richardson AL, Dillon DA, Ly A, Brock JE (2013) Interobserver concordance in implementing the 2010 ASCO/CAP recommendations for reporting ER in breast carcinomas: a demonstration of the difficulties of consistently reporting low levels of ER expression by manual quantification. Am J Clin Pathol 140(4):487–494. https://doi.org/10.1309/ajcp1rf9fuizrdpi
Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF et al (2015) Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med 373(21):2005–2014. https://doi.org/10.1056/NEJMoa1510764
Knauer M, Mook S, Rutgers EJ, Bender RA, Hauptmann M, van de Vijver MJ et al (2010) The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res Treat 120(3):655–661. https://doi.org/10.1007/s10549-010-0814-2
Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S et al (2015) Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genom 8:54. https://doi.org/10.1186/s12920-015-0129-6
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B et al (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of early breast cancer 2013. Ann Oncol 24(9):2206–2223. https://doi.org/10.1093/annonc/mdt303
Wirtz RM, Sihto H, Isola J, Heikkilä P, Kellokumpu-Lehtinen PL, Auvinen P et al (2016) Biological subtyping of early breast cancer: a study comparing RT-qPCR with immunohistochemistry. Breast Cancer Res Treat 157(3):437–446. https://doi.org/10.1007/s10549-016-3835-7
Wu NC, Wong W, Ho KE, Chu VC, Rizo A, Davenport S et al (2018) Comparison of central laboratory assessments of ER, PR, HER2, and Ki67 by IHC/FISH and the corresponding mRNAs (ESR1, PGR, ERBB2, and MKi67) by RT-qPCR on an automated, broadly deployed diagnostic platform. Breast Cancer Res Treat 172(2):327–338. https://doi.org/10.1007/s10549-018-4889-5
Varga Z, Lebeau A, Bu H, Hartmann A, Penault-Llorca F, Guerini-Rocco E et al (2017) An international reproducibility study validating quantitative determination of ERBB2, ESR1, PGR, and MKI67 mRNA in breast cancer using MammaTyper®. Breast Cancer Res 19(1):55. https://doi.org/10.1186/s13058-017-0848-z
Balduzzi A, Bagnardi V, Rotmensz N, Dellapasqua S, Montagna E, Cardillo A et al (2014) Survival outcomes in breast cancer patients with low estrogen/progesterone receptor expression. Clin Breast Cancer 14(4):258–264. https://doi.org/10.1016/j.clbc.2013.10.019
Prat A, Cheang MC, Martín M, Parker JS, Carrasco E, Caballero R et al (2013) Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol 31(2):203–209. https://doi.org/10.1200/jco.2012.43.4134
Li AQ, Zhou SL, Li M, Xu Y, Shui RH, Yu BH et al (2015) Clinicopathologic characteristics of oestrogen receptor-positive/progesterone receptor-negative/Her2-negative breast cancer according to a novel definition of negative progesterone receptor status: a large population-based study from China. PLoS ONE 10(5):e0125067. https://doi.org/10.1371/journal.pone.0125067
Doi T, Shitara K, Naito Y, Shimomura A, Fujiwara Y, Yonemori K et al (2017) Safety, pharmacokinetics, and antitumour activity of trastuzumab deruxtecan (DS-8201), a HER2-targeting antibody-drug conjugate, in patients with advanced breast and gastric or gastro-oesophageal tumours: a phase 1 dose-escalation study. Lancet Oncol 18(11):1512–1522. https://doi.org/10.1016/s1470-2045(17)30604-6
Tarantino P, Hamilton E, Tolaney SM, Cortes J, Morganti S, Ferraro E et al (2020) HER2-low breast cancer: pathological and clinical landscape. J Clin Oncol 38(17):1951–1962. https://doi.org/10.1200/jco.19.02488
de Azambuja E, Cardoso F, de Castro Jr. G, Colozza M, Mano MS, Durbecq V et al (2007) Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer 96(10):1504–1513. https://doi.org/10.1038/sj.bjc.6603756
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ (2011) Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast cancer 2011. Ann Oncol 22(8):1736–1747. https://doi.org/10.1093/annonc/mdr304
Polley MY, Leung SC, Gao D, Mastropasqua MG, Zabaglo LA, Bartlett JM et al (2015) An international study to increase concordance in Ki67 scoring. Mod Pathol 28(6):778–786. https://doi.org/10.1038/modpathol.2015.38
Leung SCY, Nielsen TO, Zabaglo LA, Arun I, Badve SS, Bane AL et al (2019) Analytical validation of a standardised scoring protocol for Ki67 immunohistochemistry on breast cancer excision whole sections: an international multicentre collaboration. Histopathology 75(2):225–235. https://doi.org/10.1111/his.13880
Janeva S, Parris TZ, Nasic S, De Lara S, Larsson K, Audisio RA et al (2021) Comparison of breast cancer surrogate subtyping using a closed-system RT-qPCR breast cancer assay and immunohistochemistry on 100 core needle biopsies with matching surgical specimens. BMC Cancer 21(1):439. https://doi.org/10.1186/s12885-021-08171-2
Acknowledgements
The authors would like to express our gratitude for those who have critically reviewed this manuscript and those who give us help during this experiment.
Funding
This work was supported by the National Key R&D Program of China (No. 2017YFC1308800), the National Natural Science Foundation of China (30900650, 81372501, 81572260, 81773299, 81701834, 81502327, 81172232, and 31430030), and the Guangdong Natural Science Foundation (2011B031800025, S2012010008378, S2012010008270, S2013010015327, 2013B021800126, 20090171120070, 9451008901002146, 2013B021800126, 2014A030313052, 2014J4100132, 2015A020214010, 2016A020215055, 201704020094, 2013B021800259, 2017B070705002, 16ykjc08, and 2015ykzd07).
Author information
Authors and Affiliations
Contributions
ZFK, LLC, and YYC were involved in the conception of the study. LHW and PL performed the RT-qPCR assays; JL, YFW, JWZ, WHZ, and HXL provided study data and materials; YYC, ZPX, LLC, HXL, JL, MS, and LLH performed the statistical analysis and wrote the statistical plan; LLC, ZPX, YYC, LHW, and PL interpreted the data; LLC, YYC, and ZPX drafted the manuscript; ZFK and LLC critically revised the article, and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflicts of interest.
Consent for publication
All authors have read and approved the version of the manuscript.
Ethical approval
Approval for the study was granted by the Ethics Committee of First Affiliated Hospital of Sun Yat-sen University (No. 2016-032). All participants signed the consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, L., Chen, Y., Xie, Z. et al. Comparison of immunohistochemistry and RT-qPCR for assessing ER, PR, HER2, and Ki67 and evaluating subtypes in patients with breast cancer. Breast Cancer Res Treat 194, 517–529 (2022). https://doi.org/10.1007/s10549-022-06649-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-022-06649-6