Abstract
Purpose
Breast cancer (BC) is considered a heterogeneous disease composed of distinct subtypes with diverse clinical outcomes. Luminal subtype tumors have the best prognosis, and patients benefit from endocrine therapy. However, resistance to endocrine therapies in BC is an obstacle to successful treatment, and novel biomarkers are needed to understand and overcome this mechanism. The RET, BCAR1, and BCAR3 genes may be associated with BC progression and endocrine resistance.
Methods
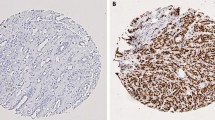
Aiming to evaluate the expression profile and prognostic value of RET, BCAR1, and BCAR3, we performed immunohistochemistry on tissue microarrays (TMAs) containing a cohort of 361 Luminal subtype BC.
Results
Low expression levels of these three proteins were predominantly observed. BCAR1 expression was correlated with nuclear grade (p = 0.057), and BCAR3 expression was correlated with lymph node status (p = 0.011) and response to hormonal therapy (p = 0.021). Further, low expression of either BCAR1 or BCAR3 was significantly associated with poor prognosis (p = 0.005; p = 0.042). Pairwise analysis showed that patients with tumors with low BCAR1/low BCAR3 expression had a poorer overall survival (p = 0.013), and the low BCAR3 expression had the worst prognosis with RET high expression stratifying these patients into two different groups. Regarding the response to hormonal therapy, non-responder patients presented lower expression of RET in comparison to the responder group (p = 0.035). Additionally, the low BCAR1 expression patients had poorer outcomes than BCAR1 high (p = 0.015).
Conclusion
Our findings suggest RET, BCAR1, and BCAR3 as potential candidate markers for endocrine therapy resistance in Luminal BC.




Similar content being viewed by others
References
Beca F, Polyak K (2016) Intratumor heterogeneity in breast cancer. Adv Exp Med Biol 882:169–189. https://doi.org/10.1007/978-3-319-22909-6_7
Braunstein LZ, Taghian AG (2016) Molecular phenotype, multigene assays, and the locoregional management of breast cancer. Semin Radiat Oncol 26:9–16. https://doi.org/10.1016/j.semradonc.2015.08.002
Mulligan LM (2014) RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer 14:173–186. https://doi.org/10.1038/nrc3680
Plaza-Menacho I, Mologni L, McDonald NQ (2014) Mechanisms of RET signaling in cancer: current and future implications for targeted therapy. Cell Signal 26:1743–1752. https://doi.org/10.1016/j.cellsig.2014.03.032
Boulay A, Breuleux M, Stephan C, Fux C, Brisken C, Fiche M, Wartmann M, Stumm M, Lane HA, Hynes NE (2008) The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res 68:3743–3751. https://doi.org/10.1158/0008-5472.CAN-07-5100
Esseghir S, Todd SK, Hunt T, Poulsom R, Plaza-Menacho I, Reis-Filho JS, Isacke CM (2007) A role for glial cell derived neurotrophic factor induced expression by inflammatory cytokines and RET/GFR alpha 1 receptor up-regulation in breast cancer. Cancer Res 67:11732–11741. https://doi.org/10.1158/0008-5472.CAN-07-2343
Gattelli A, Nalvarte I, Boulay A, Roloff TC, Schreiber M, Carragher N, Macleod KK, Schlederer M, Lienhard S, Kenner L et al (2013) Ret inhibition decreases growth and metastatic potential of estrogen receptor positive breast cancer cells. EMBO Mol Med 5:1335–1350. https://doi.org/10.1002/emmm.201302625
Morandi A, Martin LA, Gao Q, Pancholi S, Mackay A, Robertson D, Zvelebil M, Dowsett M, Plaza-Menacho I, Isacke CM (2013) GDNF-RET signaling in ER-positive breast cancers is a key determinant of response and resistance to aromatase inhibitors. Cancer Res 73:3783–3795. https://doi.org/10.1158/0008-5472.CAN-12-4265
Plaza-Menacho I, Morandi A, Robertson D, Pancholi S, Drury S, Dowsett M, Martin LA, Isacke CM (2010) Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene 29:4648–4657. https://doi.org/10.1038/onc.2010.209
Gattelli A, Hynes NE, Schor IE, Vallone SA (2020) Ret receptor has distinct alterations and functions in breast cancer. J Mammary Gland Biol Neoplasia 25:13–26. https://doi.org/10.1007/s10911-020-09445-4
Arpino G, Wiechmann L, Osborne CK, Schiff R (2008) Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev 29:217–233. https://doi.org/10.1210/er.2006-0045
Morandi A, Plaza-Menacho I, Isacke CM (2011) RET in breast cancer: functional and therapeutic implications. Trends Mol Med 17:149–157. https://doi.org/10.1016/j.molmed.2010.12.007
Mechera R, Soysal SD, Piscuoglio S, Ng CKY, Zeindler J, Mujagic E, Däster S, Glauser P, Hoffmann H, Kilic E et al (2019) Expression of RET is associated with Oestrogen receptor expression but lacks prognostic significance in breast cancer. BMC Cancer 19:41. https://doi.org/10.1186/s12885-018-5262-0
Koytiger G, Kaushansky A, Gordus A, Rush J, Sorger PK, MacBeath G (2013) Phosphotyrosine signaling proteins that drive oncogenesis tend to be highly interconnected. Mol Cell Proteomics 12:1204–1213. https://doi.org/10.1074/mcp.M112.025858
Cabodi S, Tinnirello A, Di Stefano P, Bisarò B, Ambrosino E, Castellano I, Sapino A, Arisio R, Cavallo F, Forni G et al (2006) p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res 66:4672–4680. https://doi.org/10.1158/0008-5472.CAN-05-2909
Tikhmyanova N, Little JL, Golemis EA (2010) CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci 67:1025–1048. https://doi.org/10.1007/s00018-009-0213-1
Brinkman A, van der Flier S, Kok EM, Dorssers LC (2000) BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst 92:112–120. https://doi.org/10.1093/jnci/92.2.112
van Agthoven T, van Agthoven TL, Dekker A, van der Spek PJ, Vreede L, Dorssers LC (1998) Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J 17:2799–2808. https://doi.org/10.1093/emboj/17.10.2799
Felekkis KN, Narsimhan RP, Near R, Castro AF, Zheng Y, Quilliam LA, Lerner A (2005) AND-34 activates phosphatidylinositol 3-kinase and induces anti-estrogen resistance in a SH2 and GDP exchange factor-like domain-dependent manner. Mol Cancer Res 3:32–41
Schrecengost RS, Riggins RB, Thomas KS, Guerrero MS, Bouton AH (2007) Breast cancer antiestrogen resistance-3 expression regulates breast cancer cell migration through promotion of p130Cas membrane localization and membrane ruffling. Cancer Res 67:6174–6182. https://doi.org/10.1158/0008-5472.CAN-06-3455
Cai D, Iyer A, Felekkis KN, Near RI, Luo Z, Chernoff J, Albanese C, Pestell RG, Lerner A (2003) AND-34/BCAR3, a GDP exchange factor whose overexpression confers antiestrogen resistance, activates Rac, PAK1, and the cyclin D1 promoter. Cancer Res 63:6802–6808
Cai D, Felekkis KN, Near RI, O’Neill GM, van Seventer JM, Golemis EA, Lerner A (2003) The GDP exchange factor AND-34 is expressed in B cells, associates with HEF1, and activates Cdc42. J Immunol 170:969–978. https://doi.org/10.4049/jimmunol.170.2.969
Wallez Y, Mace PD, Pasquale EB, Riedl SJ (2012) NSP-CAS protein complexes: emerging signaling modules in cancer. Genes Cancer 3:382–393. https://doi.org/10.1177/1947601912460050
Cross AM, Wilson AL, Guerrero MS, Thomas KS, Bachir AI, Kubow KE, Horwitz AR, Bouton AH (2016) Breast cancer antiestrogen resistance 3–p130. Oncogene 35:5850–5859. https://doi.org/10.1038/onc.2016.123
van Agthoven T, Godinho MF, Wulfkuhle JD, Petricoin EF, Dorssers LC (2012) Protein pathway activation mapping reveals molecular networks associated with antiestrogen resistance in breast cancer cell lines. Int J Cancer 131:1998–2007. https://doi.org/10.1002/ijc.27489
Nagai MA, Gerhard R, Fregnani JH, Nonogaki S, Rierger RB, Netto MM, Soares FA (2011) Prognostic value of NDRG1 and SPARC protein expression in breast cancer patients. Breast Cancer Res Treat 126:1–14. https://doi.org/10.1007/s10549-010-0867-2
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG et al (2017) QuPath: open source software for digital pathology image analysis. Sci Rep 7:16878. https://doi.org/10.1038/s41598-017-17204-5
Ciriello G, Sinha R, Hoadley KA, Jacobsen AS, Reva B, Perou CM, Sander C, Schultz N (2013) The molecular diversity of Luminal A breast tumors. Breast Cancer Res Treat 141:409–420. https://doi.org/10.1007/s10549-013-2699-3
Brufsky AM, Dickler MN (2018) Estrogen receptor-positive breast cancer: exploiting signaling pathways implicated in endocrine resistance. Oncologist 23:528–539. https://doi.org/10.1634/theoncologist.2017-0423
Clarke R, Tyson JJ, Dixon JM (2015) Endocrine resistance in breast cancer–an overview and update. Mol Cell Endocrinol 418(Pt 3):220–234. https://doi.org/10.1016/j.mce.2015.09.035
Spanheimer PM, Park JM, Askeland RW, Kulak MV, Woodfield GW, De Andrade JP, Cyr AR, Sugg SL, Thomas A, Weigel RJ (2014) Inhibition of RET increases the efficacy of antiestrogen and is a novel treatment strategy for luminal breast cancer. Clin Cancer Res 20:2115–2125. https://doi.org/10.1158/1078-0432.CCR-13-2221
AlFakeeh A, Brezden-Masley C (2018) Overcoming endocrine resistance in hormone receptor-positive breast cancer. Curr Oncol 25:S18–S27. https://doi.org/10.3747/co.25.3752
Drilon A, Hu ZI, Lai GGY, Tan DSW (2018) Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol 15:150. https://doi.org/10.1038/nrclinonc.2017.188
Lo Nigro C, Rusmini M, Ceccherini I (2019) RET in breast cancer: pathogenic implications and mechanisms of drug resistance. Cancer Drug Resistance 2:1136–1152. https://doi.org/10.20517/cdr.2019.66
Near RI, Zhang Y, Makkinje A, Vanden Borre P, Lerner A (2007) AND-34/BCAR3 differs from other NSP homologs in induction of anti-estrogen resistance, cyclin D1 promoter activation and altered breast cancer cell morphology. J Cell Physiol 212:655–665. https://doi.org/10.1002/jcp.21059
Wilson AL, Schrecengost RS, Guerrero MS, Thomas KS, Bouton AH (2013) Breast cancer antiestrogen resistance 3 (BCAR3) promotes cell motility by regulating actin cytoskeletal and adhesion remodeling in invasive breast cancer cells. PLoS One 8:e65678. https://doi.org/10.1371/journal.pone.0065678
Guo J, Canaff L, Rajadurai CV, Fils-Aimé N, Tian J, Dai M, Korah J, Villatoro M, Park M, Ali S et al (2014) Breast cancer anti-estrogen resistance 3 inhibits transforming growth factor β/Smad signaling and associates with favorable breast cancer disease outcomes. Breast Cancer Res 16:476. https://doi.org/10.1186/s13058-014-0476-9
van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, Look MP, Smid M, Veldscholte J, Sleijfer S, Foekens JA, Dorssers LC (2009) Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol 27:542–549. https://doi.org/10.1200/JCO.2008.17.1462
Sieuwerts AM, Lyng MB, Meijer-van Gelder ME, de Weerd V, Sweep FC, Foekens JA, Span PN, Martens JW, Ditzel HJ (2014) Evaluation of the ability of adjuvant tamoxifen-benefit gene signatures to predict outcome of hormone-naive estrogen receptor-positive breast cancer patients treated with tamoxifen in the advanced setting. Mol Oncol 8:1679–1689. https://doi.org/10.1016/j.molonc.2014.07.003
Tornillo G, Elia AR, Castellano I, Spadaro M, Bernabei P, Bisaro B, Camacho-Leal MeP, Pincini A, Provero P, Sapino A, et al (2013) p130Cas alters the differentiation potential of mammary luminal progenitors by deregulating c-Kit activity. Stem Cells 31: 1422–1433. https://doi.org/10.1002/stem.1403.
Tornillo G, Defilippi P, Cabodi S (2014) Cas proteins: dodgy scaffolding in breast cancer. Breast Cancer Res 16:443. https://doi.org/10.1186/s13058-014-0443-5
van der Flier S, Brinkman A, Look MP, Kok EM, Meijer-van Gelder ME, Klijn JG, Dorssers LC, Foekens JA (2000) Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst 92:120–127. https://doi.org/10.1093/jnci/92.2.120
Subbiah V, Yang D, Velcheti V, Drilon A, Meric-Bernstam F (2020) State-of-the-art strategies for targeting. J Clin Oncol 38:1209–1221. https://doi.org/10.1200/JCO.19.02551
Funding
This work was supported by the Fundação de Amparo a Pesquisa do Estadode São Paulo (FAPESP; 2019/05252-4) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 309524/2017-2).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Ethical approval
This study was conducted according to the guidelines laid down in the Declaration of Helsinki and was approved by the Institutional Review Boards of the Medical School University of São Paulo, São Paulo, Brasil (CAAE: 35295514.1.0000.0065).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10549_2021_6452_MOESM1_ESM.tiff
Supplementary file1—Supplementary Figure 1 - Kaplan-Meier curves of disease-free survival in Luminal breast cancer patients stratified according to RET (a), BCAR1 (b), and BCAR3 (c) protein expression. Tumors were classified as low (negative or weak) or high (moderate or intense) according to RET, BCAR1, or BCAR3 protein immunostaining. Log-rank test was performed for curve comparisons. HR - hazard ratio. (TIFF 109 kb)
10549_2021_6452_MOESM2_ESM.tiff
Supplementary file2—Supplementary Figure 2 - Prognostic association of the pairwise combinations of RET, BCAR1 and BCAR3 expression. Kaplan-Meier curves of disease-free survival in luminal breast cancer patients stratified according to the combination of RET and BCAR3 (a), RET and BCAR1 (b) and, BCAR3 and BCAR1 (c) protein expression. Tumors were classified as low (negative or weak) or high (moderate or intense) according to RET, BCAR3, or BCAR1 protein immunostaining. Log-rank test was performed for curve comparisons. NA – not analyzed because 100% of the cases was censored cases. HR -hazard ratio. (TIFF 331 kb)
10549_2021_6452_MOESM3_ESM.tiff
Supplementary file3—Supplementary Figure 3 - Prognostic association between RET, BCAR1, and BCAR3 expression and response to endocrine therapy. Kaplan-Meier curves of disease-free survival in Luminal breast cancer patients stratified according to RET (a), BCAR1 (b), and BCAR3 (c) protein expression in combination with response to endocrine therapy. Tumors were classified as low (negative or weak) or high (moderate or intense) according to RET, BCAR1, or BCAR3 protein immunostaining and response to hormonal therapy was considered as absence of disease recurrence. Log-rank test was performed for curve comparisons. The p-values obtained for comparison between responder and/or non responder curves are presented inside the figures. NA – not analyzed because 100% of the cases was censored cases. HR - Hazard ratio. (TIFF 127 kb)
Rights and permissions
About this article
Cite this article
Pavanelli, A.C., Mangone, F.R., Yoganathan, P. et al. Comprehensive immunohistochemical analysis of RET, BCAR1, and BCAR3 expression in patients with Luminal A and B breast cancer subtypes. Breast Cancer Res Treat 192, 43–52 (2022). https://doi.org/10.1007/s10549-021-06452-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-021-06452-9