Abstract
Purpose
Patients with triple-negative breast cancer (TNBC) who do not achieve pathological complete response (pCR) following neoadjuvant chemotherapy have a high risk of recurrence and death. Molecular characterization may identify patients unlikely to achieve pCR. This neoadjuvant trial was conducted to determine the pCR rate with docetaxel and carboplatin and to identify molecular alterations and/or immune gene signatures predicting pCR.
Experimental design
Patients with clinical stages II/III TNBC received 6 cycles of docetaxel and carboplatin. The primary objective was to determine if neoadjuvant docetaxel and carboplatin would increase the pCR rate in TNBC compared to historical expectations. We performed whole-exome sequencing (WES) and immune profiling on pre-treatment tumor samples to identify alterations that may predict pCR. Thirteen matching on-treatment samples were also analyzed to assess changes in molecular profiles.
Results
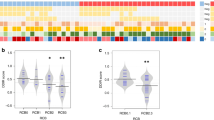
Fifty-eight of 127 (45.7%) patients achieved pCR. There was a non-significant trend toward higher mutation burden for patients with residual cancer burden (RCB) 0/I versus RCB II/III (median 80 versus 68 variants, p 0.88). TP53 was the most frequently mutated gene, observed in 85.7% of tumors. EGFR, RB1, RAD51AP2, SDK2, L1CAM, KPRP, PCDHA1, CACNA1S, CFAP58, COL22A1, and COL4A5 mutations were observed almost exclusively in pre-treatment samples from patients who achieved pCR. Seven mutations in PCDHA1 were observed in pre-treatment samples from patients who did not achieve pCR. Several immune gene signatures including IDO1, PD-L1, interferon gamma signaling, CTLA4, cytotoxicity, tumor inflammation signature, inflammatory chemokines, cytotoxic cells, lymphoid, PD-L2, exhausted CD8, Tregs, and immunoproteasome were upregulated in pre-treatment samples from patients who achieved pCR.
Conclusion
Neoadjuvant docetaxel and carboplatin resulted in a pCR of 45.7%. WES and immune profiling differentiated patients with and without pCR.
Trial registration: Clinical trial information: NCT02124902, Registered 24 April 2014 & NCT02547987, Registered 10 September 2015.






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Abbreviations
- TNBC:
-
Triple-negative breast cancer
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
HER2/Neu gene
- DNA:
-
Deoxyribonucleic acid
- pCR:
-
Pathological complete response
- IHC:
-
Immunohistochemistry
- FISH:
-
Fluorescence in situ hybridization
- WUSM:
-
Washington University School of Medicine
- BCM:
-
Baylor College of Medicine
- AUC:
-
Area under the curve
- NCI CTCAE:
-
National Cancer Institute Common Terminology Criteria for Adverse Events
- C1D3:
-
Cycle 1 day 3
- RCB:
-
Residual cancer burden
- TIL:
-
Tumor-infiltrating lymphocytes
- WES:
-
Whole-exome sequencing
- SNV:
-
Single-nucleotide variant
- RNA:
-
Ribonucleic acid
- FFPE:
-
Formalin-fixed paraffin-embedded
- TIS:
-
Tumor inflammation signature
- IUO:
-
Investigational use only
- RUO:
-
Research use only
- SKAT:
-
Sequence kernel-based association
- SAE:
-
Serious adverse events
- ALC:
-
Absolute lymphocyte count
- OR:
-
Odds ratio
- VAF:
-
Variant allele frequency
- TME:
-
Tumor microenvironment
References
DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL (2019) Breast cancer statistics. CA Cancer J Clin 69(6):438–451. https://doi.org/10.3322/caac.21583
Masuda N, Lee SJ, Ohtani S, Im YH, Lee ES, Yokota I, Kuroi K, Im SA, Park BW, Kim SB, Yanagita Y, Ohno S, Takao S, Aogi K, Iwata H, Jeong J, Kim A, Park KH, Sasano H, Ohashi Y, Toi M (2017) Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N Engl J Med 376(22):2147–2159. https://doi.org/10.1056/NEJMoa1612645
Early Breast Cancer Trialists’ Collaborative G, Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, Cutter D, Darby S, McGale P, Taylor C, Wang YC, Bergh J, Di Leo A, Albain K, Swain S, Piccart M, Pritchard K (2012) Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet 379(9814):432–444. https://doi.org/10.1016/S0140-6736(11)61625-5
Henderson IC, Berry DA, Demetri GD, Cirrincione CT, Goldstein LJ, Martino S, Ingle JN, Cooper MR, Hayes DF, Tkaczuk KH, Fleming G, Holland JF, Duggan DB, Carpenter JT, Frei E 3rd, Schilsky RL, Wood WC, Muss HB, Norton L (2003) Improved outcomes from adding sequential Paclitaxel but not from escalating Doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol 21(6):976–983. https://doi.org/10.1200/JCO.2003.02.063
Byrski T, Dent R, Blecharz P, Foszczynska-Kloda M, Gronwald J, Huzarski T, Cybulski C, Marczyk E, Chrzan R, Eisen A, Lubinski J, Narod SA (2012) Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res 14(4):R110. https://doi.org/10.1186/bcr3231
Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, Kladny J, Gorski B, Lubinski J, Narod SA (2009) Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 115(2):359–363. https://doi.org/10.1007/s10549-008-0128-9
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100(14):8418–8423. https://doi.org/10.1073/pnas.0932692100
Kern P, Kalisch A, Kolberg HC, Kimmig R, Otterbach F, von Minckwitz G, Sikov WM, Pott D, Kurbacher C (2013) Neoadjuvant, anthracycline-free chemotherapy with carboplatin and docetaxel in triple-negative, early-stage breast cancer: a multicentric analysis of feasibility and rates of pathologic complete response. Chemotherapy 59(5):387–394. https://doi.org/10.1159/000362756
Kern P, Kalisch A, von Minckwitz G, Putter C, Kolberg HC, Pott D, Kurbacher C, Rezai M, Kimmig R (2016) Neoadjuvant, anthracycline-free chemotherapy with carboplatin and docetaxel in triple-negative, early-stage breast cancer: a multicentric analysis of rates of pathologic complete response and survival. J Chemother 28(3):210–217. https://doi.org/10.1179/1973947815Y.0000000061
Sharma P, Lopez-Tarruella S, Garcia-Saenz JA, Khan QJ, Gomez HL, Prat A, Moreno F, Jerez-Gilarranz Y, Barnadas A, Picornell AC, Monte-Millan MD, Gonzalez-Rivera M, Massarrah T, Pelaez-Lorenzo B, Palomero MI, Gonzalez Del Val R, Cortes J, Fuentes-Rivera H, Morales DB, Marquez-Rodas I, Perou CM, Lehn C, Wang YY, Klemp JR, Mammen JV, Wagner JL, Amin AL, O’Dea AP, Heldstab J, Jensen RA, Kimler BF, Godwin AK, Martin M (2018) Pathological response and survival in triple-negative breast cancer following neoadjuvant carboplatin plus Docetaxel. Clin Cancer Res 24(23):5820–5829. https://doi.org/10.1158/1078-0432.CCR-18-0585
Sharma P, Lopez-Tarruella S, Garcia-Saenz JA, Ward C, Connor CS, Gomez HL, Prat A, Moreno F, Jerez-Gilarranz Y, Barnadas A, Picornell AC, Del Monte-Millan M, Gonzalez-Rivera M, Massarrah T, Pelaez-Lorenzo B, Palomero MI, Gonzalez Del Val R, Cortes J, Fuentes Rivera H, Bretel Morales D, Marquez-Rodas I, Perou CM, Wagner JL, Mammen JM, McGinness MK, Klemp JR, Amin AL, Fabian CJ, Heldstab J, Godwin AK, Jensen RA, Kimler BF, Khan QJ, Martin M (2017) Efficacy of neoadjuvant carboplatin plus Docetaxel in triple-negative breast cancer: combined analysis of two cohorts. Clin Cancer Res 23(3):649–657. https://doi.org/10.1158/1078-0432.CCR-16-0162
Aparicio S, Caldas C (2013) The implications of clonal genome evolution for cancer medicine. N Engl J Med 368(9):842–851. https://doi.org/10.1056/NEJMra1204892
Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Kuzma CS, Pluard TJ, Somlo G, Port ER, Golshan M, Bellon JR, Collyar D, Hahn OM, Carey LA, Hudis CA, Winer EP (2015) Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 33(1):13–21. https://doi.org/10.1200/JCO.2014.57.0572
von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, Blohmer JU, Jackisch C, Paepke S, Gerber B, Zahm DM, Kummel S, Eidtmann H, Klare P, Huober J, Costa S, Tesch H, Hanusch C, Hilfrich J, Khandan F, Fasching PA, Sinn BV, Engels K, Mehta K, Nekljudova V, Untch M (2014) Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol 15(7):747–756. https://doi.org/10.1016/S1470-2045(14)70160-3
Gerber B, Loibl S, Eidtmann H, Rezai M, Fasching PA, Tesch H, Eggemann H, Schrader I, Kittel K, Hanusch C, Kreienberg R, Solbach C, Jackisch C, Kunz G, Blohmer JU, Huober J, Hauschild M, Nekljudova V, Untch M, von Minckwitz G, German Breast Group I (2013) Neoadjuvant bevacizumab and anthracycline-taxane-based chemotherapy in 678 triple-negative primary breast cancers; results from the geparquinto study (GBG 44). Ann Oncol 24(12):2978–2984. https://doi.org/10.1093/annonc/mdt361
von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Konecny GE, Denkert C, Nekljudova V, Mehta K, Loibl S (2012) Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 30(15):1796–1804. https://doi.org/10.1200/JCO.2011.38.8595
Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26(8):1275–1281. https://doi.org/10.1200/JCO.2007.14.4147
Vargo JA, Beriwal S, Ahrendt GM, Soran A, Johnson RR, McGuire K, Bhargava R (2011) Molecular class as a predictor of locoregional and distant recurrence in the neoadjuvant setting for breast cancer. Oncology 80(5–6):341–349. https://doi.org/10.1159/000330203
Wang S, Yang H, Tong F, Zhang J, Yang D, Liu H, Cao Y, Liu P, Zhou P, Cheng L, Liu M, Guo J (2009) Response to neoadjuvant therapy and disease free survival in patients with triple-negative breast cancer. Gan To Kagaku Ryoho 36(2):255–258
Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM (2007) The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13(8):2329–2334. https://doi.org/10.1158/1078-0432.ccr-06-1109
Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Somlo G, Port ER, Qamar R, Sturtz K, Mamounas E, Golshan M, Bellon JR, Collyar D, Hahn OM, Carey LA, Hudis CA, Winer EP (2015) Event-free and overall survival following neoadjuvant weekly paclitaxel and dose-dense AC +/− carboplatin and/or bevacizumab in triple-negative breast cancer: outcomes from CALGB 40603 (Alliance). Paper presented at the San Antonio Breast Cancer Symposium, San Antonio
Geiger S, Cnossen JA, Horster S, DiGioia D, Heinemann V, Stemmler HJ (2011) Long-term follow-up of patients with metastatic breast cancer: results of a retrospective, single-center analysis from 2000 to 2005. Anticancer Drugs 22(9):933–939. https://doi.org/10.1097/CAD.0b013e32834860af
Kassam F, Enright K, Dent R, Dranitsaris G, Myers J, Flynn C, Fralick M, Kumar R, Clemons M (2009) Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clin Breast Cancer 9(1):29–33. https://doi.org/10.3816/CBC.2009.n.005
Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Dieras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA, Investigators IMT (2018) Atezolizumab and Nab-Paclitaxel in advanced triple-negative breast cancer. N Engl J Med 379(22):2108–2121. https://doi.org/10.1056/NEJMoa1809615
Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, Perez EA, Thompson EA, Symmans WF, Richardson AL, Brock J, Criscitiello C, Bailey H, Ignatiadis M, Floris G, Sparano J, Kos Z, Nielsen T, Rimm DL, Allison KH, Reis-Filho JS, Loibl S, Sotiriou C, Viale G, Badve S, Adams S, Willard-Gallo K, Loi S, International TWG (2015) The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 26(2):259–271. https://doi.org/10.1093/annonc/mdu450
Voss K, Van der Auwera G, Gentry J (2017) Full-stack genomics pipelining with GATK4 + WDL + Cromwell. In: 18th Annual Bioinformatics Open Source Conference, Prague, Czech Republic, 2017
Griffith M, Griffith OL, Smith SM, Ramu A, Callaway MB, Brummett AM, Kiwala MJ, Coffman AC, Regier AA, Oberkfell BJ, Sanderson GE, Mooney TP, Nutter NG, Belter EA, Du F, Long RL, Abbott TE, Ferguson IT, Morton DL, Burnett MM, Weible JV, Peck JB, Dukes A, McMichael JF, Lolofie JT, Derickson BR, Hundal J, Skidmore ZL, Ainscough BJ, Dees ND, Schierding WS, Kandoth C, Kim KH, Lu C, Harris CC, Maher N, Maher CA, Magrini VJ, Abbott BS, Chen K, Clark E, Das I, Fan X, Hawkins AE, Hepler TG, Wylie TN, Leonard SM, Schroeder WE, Shi X, Carmichael LK, Weil MR, Wohlstadter RW, Stiehr G, McLellan MD, Pohl CS, Miller CA, Koboldt DC, Walker JR, Eldred JM, Larson DE, Dooling DJ, Ding L, Mardis ER, Wilson RK (2015) Genome modeling system: a knowledge management platform for genomics. PLoS Comput Biol 11(7):e1004274. https://doi.org/10.1371/journal.pcbi.1004274
Li H (2020) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. https://arxiv.org/abs/1303.3997. Accessed 20 July 2020
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. https://doi.org/10.1101/gr.107524.110
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
do Valle IF, Giampieri E, Simonetti G, Padella A, Manfrini M, Ferrari A, Papayannidis C, Zironi I, Garonzi M, Bernardi S, Delledonne M, Martinelli G, Remondini D, Castellani G (2016) Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinform 17(Suppl 12):341. https://doi.org/10.1186/s12859-016-1190-7
Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK (2012) Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28(14):1811–1817. https://doi.org/10.1093/bioinformatics/bts271
Koboldt DC, Larson DE, Wilson RK (2013) Using VarScan 2 for germline variant calling and somatic mutation detection. Curr Protoc Bioinform. https://doi.org/10.1002/0471250953.bi1504s44
Tan A, Abecasis GR, Kang HM (2015) Unified representation of genetic variants. Bioinformatics 31(13):2202–2204. https://doi.org/10.1093/bioinformatics/btv112
Karczewski K, Francioli L, Tiao G, Cummings B, Alföldi J, Wang Q, Collins R, Laricchia K, Ganna A, Birnbaum D, Gauthier L, Brand H, Solomonson M, Watts N, Rhodes D, Singer-Berk M, Seaby E, Kosmicki J, Walters R, Macarthur D (2019) Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes: supplementary information. https://doi.org/10.1101/531210
Barnell EK, Ronning P, Campbell KM, Krysiak K, Ainscough BJ, Sheta LM, Pema SP, Schmidt AD, Richters M, Cotto KC, Danos AM, Ramirez C, Skidmore ZL, Spies NC, Hundal J, Sediqzad MS, Kunisaki J, Gomez F, Trani L, Matlock M, Wagner AH, Swamidass SJ, Griffith M, Griffith OL (2019) Standard operating procedure for somatic variant refinement of sequencing data with paired tumor and normal samples. Genet Med 21(4):972–981. https://doi.org/10.1038/s41436-018-0278-z
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F (2016) The ensembl variant effect predictor. Genome Biol 17(1):122. https://doi.org/10.1186/s13059-016-0974-4
Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Marugan JC, Cummins C, Davidson C, Dodiya K, Fatima R, Gall A, Giron CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Lavidas I, Le T, Lemos D, Martinez JG, Maurel T, McDowall M, McMahon A, Mohanan S, Moore B, Nuhn M, Oheh DN, Parker A, Parton A, Patricio M, Sakthivel MP, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sycheva M, Szuba M, Taylor K, Thormann A, Threadgold G, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Flint B, Frankish A, Hunt SE, G II, Kostadima M, Langridge N, Loveland JE, Martin FJ, Morales J, Mudge JM, Muffato M, Perry E, Ruffier M, Trevanion SJ, Cunningham F, Howe KL, Zerbino DR, Flicek P (2020) Ensembl 2020. Nucleic Acids Res 48(D1):D682–D688. https://doi.org/10.1093/nar/gkz966
Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B (2019) WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47(W1):W199-w205. https://doi.org/10.1093/nar/gkz401
Damotte D, Warren S, Arrondeau J, Boudou-Rouquette P, Mansuet-Lupo A, Biton J, Ouakrim H, Alifano M, Gervais C, Bellesoeur A, Kramkimel N, Tlemsani C, Burroni B, Duche A, Letourneur F, Si H, Halpin R, Creasy T, Herbst R, Ren X, Morel P, Cesano A, Goldwasser F, Leroy K (2019) The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J Transl Med 17(1):357. https://doi.org/10.1186/s12967-019-2100-3
Vadakekolathu J, Minden MD, Hood T, Church SE, Reeder S, Altmann H, Sullivan AH, Viboch EJ, Patel T, Ibrahimova N, Warren SE, Arruda A, Liang Y, Smith TH, Foulds GA, Bailey MD, Gowen-MacDonald J, Muth J, Schmitz M, Cesano A, Pockley AG, Valk PJM, Lowenberg B, Bornhauser M, Tasian SK, Rettig MP, Davidson-Moncada JK, DiPersio JF, Rutella S (2020) Immune landscapes predict chemotherapy resistance and immunotherapy response in acute myeloid leukemia. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aaz0463
Sharma P, Kimler BF, O'Dea A, Nye EL, Wang YY, Yoder R, Prochaska LH, Wagner JL, Amin AL, Larson K, Balanoff C, Elia M, Crane GJ, Madhusudhana S, Hoffmann MS, Sheehan M, Rodriguez RR, Jensen RA, Godwin AK, Khan QJ (2019) Results of randomized phase II trial of neoadjuvant carboplatin plus docetaxel or carboplatin plus paclitaxel followed by AC in stage I-III triple-negative breast cancer (NCT02413320). American Society of Clinical Oncology, Chicago, IL
Loibl S, Weber KE, Timms KM, Elkin EP, Hahnen E, Fasching PA, Lederer B, Denkert C, Schneeweiss A, Braun S, Salat CT, Rezai M, Blohmer JU, Zahm DM, Jackisch C, Gerber B, Klare P, Kummel S, Schem C, Paepke S, Schmutzler R, Rhiem K, Penn S, Reid J, Nekljudova V, Hartman AR, von Minckwitz G, Untch M (2018) Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response-final results from GeparSixto. Ann Oncol 29(12):2341–2347. https://doi.org/10.1093/annonc/mdy460
Hahnen E, Lederer B, Hauke J, Loibl S, Krober S, Schneeweiss A, Denkert C, Fasching PA, Blohmer JU, Jackisch C, Paepke S, Gerber B, Kummel S, Schem C, Neidhardt G, Huober J, Rhiem K, Costa S, Altmuller J, Hanusch C, Thiele H, Muller V, Nurnberg P, Karn T, Nekljudova V, Untch M, von Minckwitz G, Schmutzler RK (2017) Germline mutation status, pathological complete response, and disease-free survival in triple-negative breast cancer: secondary analysis of the GeparSixto Randomized Clinical Trial. JAMA Oncol 3(10):1378–1385. https://doi.org/10.1001/jamaoncol.2017.1007
Sikov WM, Polley M, Twohy E, Perou CM, Singh B, Berry DA, Tolaney SM, Somlo G, Port ER, Ma CX, Kuzma CS, Mamounas EP, Golshan M, Bellon JR, Collyar D, Hahn OM, Hudis CA, Winer EP, Partridge AH, Carey LA (2019) CALGB (Alliance) 40603: long-term outcomes (LTOs) after neoadjuvant chemotherapy (NACT) +/− carboplatin (Cb) and bevacizumab (Bev) in triple-negative breast cancer (TNBC). American Society of Clinical Oncology Annual Meeting 2019, Chicago, IL
Doxorubicin Hydrochloride and Cyclophosphamide Followed by Paclitaxel With or Without Carboplatin in Treating Patients With Triple-Negative Breast Cancer NCT02488967. https://clinicaltrials.gov/ct2/show/NCT02488967. Accessed 22 Feb 2020
Adjuvant Treatment of EC Followed by Taxane +/− Carboplatin in Triple-Negative Breast Cancer (TCTN) NCT02455141. https://clinicaltrials.gov/ct2/show/NCT02455141. Accessed 22 Feb 2020
Carboplatin in EARLY Triple Negative Breast Cancer Trial (PEARLY Trial) NCT02441933. https://clinicaltrials.gov/ct2/show/NCT02441933. Accessed 22 Feb 2020
Cortazar P, Geyer CE Jr (2015) Pathological complete response in neoadjuvant treatment of breast cancer. Ann Surg Oncol 22(5):1441–1446. https://doi.org/10.1245/s10434-015-4404-8
Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L, Valagussa P, Swain SM, Prowell T, Loibl S, Wickerham DL, Bogaerts J, Baselga J, Perou C, Blumenthal G, Blohmer J, Mamounas EP, Bergh J, Semiglazov V, Justice R, Eidtmann H, Paik S, Piccart M, Sridhara R, Fasching PA, Slaets L, Tang S, Gerber B, Geyer CE Jr, Pazdur R, Ditsch N, Rastogi P, Eiermann W, von Minckwitz G (2014) Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384(9938):164–172. https://doi.org/10.1016/S0140-6736(13)62422-8
Symmans WF, Wei C, Gould R, Yu X, Zhang Y, Liu M, Walls A, Bousamra A, Ramineni M, Sinn B, Hunt K, Buchholz TA, Valero V, Buzdar AU, Yang W, Brewster AM, Moulder S, Pusztai L, Hatzis C, Hortobagyi GN (2017) Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol 35(10):1049–1060. https://doi.org/10.1200/JCO.2015.63.1010
Mayer IA, Zhao F, Arteaga CL, Symmans WF, Park BH, Burnett BL, Tevaarwerk AJ, Garcia SF, Smith KL, Makower DF, Block M, Morley KA, Jani CR, Mescher C, Dewani SJ, Tawfik B, Flaum LE, Mayer EL, Sikov WM, Rodler ET, Wagner LI, DeMichele AM, Sparano JA, Wolff AC, Miller KD (2021) A randomized phase III post-operative trial of platinum-based chemotherapy vs. capecitabine in patients with residual triple-negative breast cancer (TNBC) following neoadjuvant chemotherapy: ECOG-ACRIN EA1131. American Society of Clinical Oncology Annual Meeting, 2021
Tung N, Arun B, Hacker MR, Hofstatter E, Toppmeyer DL, Isakoff SJ, Borges V, Legare RD, Isaacs C, Wolff AC, Marcom PK, Mayer EL, Lange PB, Goss AJ, Jenkins C, Krop IE, Winer EP, Schnitt SJ, Garber JE (2020) TBCRC 031: randomized phase II study of neoadjuvant cisplatin versus doxorubicin-cyclophosphamide in germline BRCA carriers with HER2-negative breast cancer (the INFORM trial). J Clin Oncol 38(14):1539–1548. https://doi.org/10.1200/JCO.19.03292
Loibl S, O’Shaughnessy J, Untch M, Sikov WM, Rugo HS, McKee MD, Huober J, Golshan M, von Minckwitz G, Maag D, Sullivan D, Wolmark N, McIntyre K, Ponce Lorenzo JJ, Metzger Filho O, Rastogi P, Symmans WF, Liu X, Geyer CE Jr (2018) Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol 19(4):497–509. https://doi.org/10.1016/S1470-2045(18)30111-6
Gluz O, Nitz U, Liedtke C, Christgen M, Grischke EM, Forstbauer H, Braun M, Warm M, Hackmann J, Uleer C, Aktas B, Schumacher C, Bangemann N, Lindner C, Kuemmel S, Clemens M, Potenberg J, Staib P, Kohls A, von Schumann R, Kates R, Kates R, Schumacher J, Wuerstlein R, Kreipe HH, Harbeck N (2018) Comparison of neoadjuvant Nab-Paclitaxel+Carboplatin vs Nab-Paclitaxel+Gemcitabine in triple-negative breast cancer: randomized WSG-ADAPT-TN Trial Results. J Natl Cancer Inst 110(6):628–637. https://doi.org/10.1093/jnci/djx258
Groheux D, Biard L, Lehmann-Che J, Teixeira L, Bouhidel FA, Poirot B, Bertheau P, Merlet P, Espie M, Resche-Rigon M, Sotiriou C, de Cremoux P (2018) Tumor metabolism assessed by FDG-PET/CT and tumor proliferation assessed by genomic grade index to predict response to neoadjuvant chemotherapy in triple negative breast cancer. Eur J Nucl Med Mol Imaging 45(8):1279–1288. https://doi.org/10.1007/s00259-018-3998-z
Riva F, Bidard FC, Houy A, Saliou A, Madic J, Rampanou A, Hego C, Milder M, Cottu P, Sablin MP, Vincent-Salomon A, Lantz O, Stern MH, Proudhon C, Pierga JY (2017) Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem 63(3):691–699. https://doi.org/10.1373/clinchem.2016.262337
Chen YH, Hancock BA, Solzak JP, Brinza D, Scafe C, Miller KD, Radovich M (2017) Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 3:24. https://doi.org/10.1038/s41523-017-0028-4
Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, Wong NS, Yip GW, Bay BH, Tan PH (2011) Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res 13(2):R35. https://doi.org/10.1186/bcr2857
Cancer Genome Atlas N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70. https://doi.org/10.1038/nature11412
Lee SW, Choi D, Heo M, Shin EC, Park SH, Kim SJ, Oh YK, Lee BH, Yang SH, Sung YC, Lee H (2020) hIL-7-hyFc, a long-acting IL-7, increased absolute lymphocyte count in healthy subjects. Clin Transl Sci 13(6):1161–1169. https://doi.org/10.1111/cts.12800
Kim JH, Kim YM, Choi D, Jo SB, Park HW, Hong SW, Park S, Kim S, Moon S, You G, Kang YW, Park Y, Lee BH, Lee SW (2020) Hybrid Fc-fused interleukin-7 induces an inflamed tumor microenvironment and improves the efficacy of cancer immunotherapy. Clin Transl Immunol 9(9):e1168. https://doi.org/10.1002/cti2.1168
Acknowledgements
Tracy Summa, Isabella Grigsby, Christina Robinson, and Kristen Otte for providing clinical research support.
Funding
FOA—Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number K12 CA167540. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Research reported in this publication was supported by NeoImmuneTech, Inc. MJE—Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U01 CA214125-03. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Research reported in this publication was supported by the Cancer Prevention & Research Institute of Texas Established Investigator Award RR140033. The content is solely the responsibility of the authors and does not necessarily represent the official views of the CPRIT. OLG—Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number K22CA188163. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. MG—Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under Award Number R00HG007940. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
FOA, IC, MJE, WG were involved in the conception and design of the study. FOA, MFR, CXM, KW, RS, LLP, RB, NB, CER, AF, TPR, LFH, AR,KC, MO participated in the acquisition of data (acquired and managed patients, provided facilities, etc.). FOA, IC, JL, MFR, ISH, BF, GJ, ZLS, AB, MR, CXM, KW, JD, LLP, RB, MO, OA, BHL, SF, SC, MA, FG, WG, OLG, MG provided analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis). All authors were involved in writing, reviewing, and/or revision of the manuscript. FOA, BF, GJ, ZLS, AB, MR, SC, MA, OLG, MG provided administrative, technical, or material support (i.e., reporting or organizing data, constructing databases). The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
FOA reports consulting for Eisai, Immunomedics, AstraZeneca, Athenex, Cardinal Health, Pfizer, AbbVie, Best Doctors, and Advance Medical. FOA reports contracted research for Immunomedics, Pfizer, Seattle Genetics, NeoImmuneTech, RNA Diagnostics, and Astellas. MFR reports consulting for Genentech, MacroGenics, Daiichi, Seattle Genetics, and Novartis. MFR reports contracted research for Pfizer. RB consulting for Genentech. RB reports contracted research for Puma Biotechnology, Inc.
Ethical Approval
The protocol and informed consent documents were approved by WUSM and BCM. Upon approval, all participating institutions agreed to follow the Declaration of Helsinki, good clinical practice guidelines, and the applicable parts of the U.S. Code of Federal Regulations.
Informed consent
Written informed consent was required for enrollment.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10549_2021_6307_MOESM1_ESM.docx
Supplementary file1 (DOCX 61 kb) Additional file 1. Study Consort Diagram. Flow diagram of clinical trial including all patients screened, registered, received intervention, and are evaluable for the primary endpoint. Also indicates number of patients from both sites. WUSM, Washington University School of Medicine and BCM, Baylor College of Medicine.
10549_2021_6307_MOESM2_ESM.docx
Supplementary file2 (DOCX 338 kb) Additional file 2. Complete linkage clustering of IO360 signatures and additional gene signatures (PIK3R2, PIK3R1, JAK1, JAK3, IL2RG, STAT3, FYN) and individual baseline samples z-scaled transformed and trimmed to plus or minus 3 (N = 66 patients).
10549_2021_6307_MOESM3_ESM.docx
Supplementary file3 (DOCX 13 kb) Additional file 3. Table of fold change and adjusted p values for immune gene signatures significantly higher at baseline versus on-treatment.
10549_2021_6307_MOESM4_ESM.docx
Supplementary file4 (DOCX 257 kb) Additional file 4. The TP53 mutation profile of patients in this study is shown in the top panel, and the TP53 mutation profile from the COSMIC database is shown in the bottom panel. Each distinct mutation is represented by a disc sized in proportion to number of affected samples and filled with the color representing its mutation class. The TP53 protein is displayed as a ruler with color-banding denoting the protein domains and numerical markings denoting the number of amino acid residues from N-terminus to C-terminus. Forty-one unique TP53 mutations were observed in 48 of 56 patients’ samples sequenced on this study.
10549_2021_6307_MOESM5_ESM.docx
Supplementary file5 (DOCX 214 kb) Additional file 5. The EGFR mutation profile of patients in our study is shown in the top panel, and the EGFR mutation profile from the COSMIC database is shown in the bottom panel. Each distinct mutation is represented by a disc sized in proportion to the number of affected samples and filled with the color representing its mutation class. The EGFR protein is displayed as a ruler with color-banding denoting the protein domains and numerical markings denoting the number of amino acid residues from N-terminus to C-terminus. Four unique EGFR mutations were observed in 3 patient samples on our study.
Rights and permissions
About this article
Cite this article
Ademuyiwa, F.O., Chen, I., Luo, J. et al. Immunogenomic profiling and pathological response results from a clinical trial of docetaxel and carboplatin in triple-negative breast cancer. Breast Cancer Res Treat 189, 187–202 (2021). https://doi.org/10.1007/s10549-021-06307-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-021-06307-3