Abstract
Purpose
Estrogen receptor positive (ER+) breast cancer constitutes almost 85% of all breast cancer patients and are a genetically highly heterogenic group. Data on the association of somatic alterations to outcome and prognosis are however sparse. In this neoadjuvant endocrine phase II trial including postmenopausal breast cancer patients with ER+, HER2 normal breast cancer, we investigated the rate of pathogenic mutations before and after treatment as well as the association with treatment response and survival.
Methods
Pretreatment and posttreatment tumour samples from 109 patients treated with neoadjuvant letrozole were collected and analysed with Next Generation Sequencing utilizing a panel of 12 genes (ALK, BRAF, EGFR, ERBB2, ERBB3, ESR1, KIT, KRAS, NRAS, PDGFRA, PIK3CA, and RAF1). Residual disease was assessed by a modified Miller Payne scale and the Residual Cancer Burden index. Survival data were collected prospectively.
Results
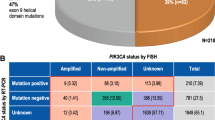
Among the 109 patients, 52 had at least one pathogenic mutation in the pretreatment sample and 60 in the posttreatment sample. The most frequently mutated gene was PIK3CA, followed by EGFR and KRAS. Twelve different pathogenic PIK3CA mutations were identified, primarily in exon 20 and exon 9. An altered PIK3CA mutation profile from the pre- to the posttreatment specimen was significantly associated to improved pathological outcome. Overall and Disease-Free Survival benefits in PIK3CA mutated patients was observed.
Conclusion
Considerable heterogeneity was identified both among patients and between pre- and posttreatment samples. PIK3CA has the potential to be a predictive biomarker. To further assess the implications of a treatment related altered PIK3CA mutation profile, more data are needed.


Similar content being viewed by others
Data availability
The data supporting all the figures, tables and supplementary tables in the published article, are not publicly available due to institutional restrictions. The dataset can be made available to qualified researchers through application to the Danish Breast Cancer Group. Please contact dbcg.rigshospitalet@regionh.dk.
References
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70
Luen SJ, Asher R, Lee CK, Savas P, Kammler R, Dell’Orto P et al (2018) Association of somatic driver alterations with prognosis in postmenopausal, hormone receptor-positive, HER2-negative early breast cancer: a secondary analysis of the BIG 1–98 randomized clinical trial. JAMA Oncol 4(10):1335–1343
Griffith OL, Spies NC, Anurag M, Griffith M, Luo J, Tu D et al (2018) The prognostic effects of somatic mutations in ER-positive breast cancer. Nature Commun 9(1):1–16
Selli C, Turnbull AK, Pearce DA, Li A, Fernando A, Wills J, et al (2019) Molecular changes during extended neoadjuvant letrozole treatment of breast cancer: distinguishing acquired resistance from dormant tumours. Breast Cancer Res 21(1):2.
Dunbier AK, Ghazoui Z, Anderson H, Salter J, Nerurkar A, Osin P et al (2013) Molecular profiling of aromatase inhibitor-treated postmenopausal breast tumors identifies immune-related correlates of resistance. Clin Cancer Res 19(10):2775–2786
Miller WR, Larionov A, Anderson TJ, Evans DB, Dixon JM (2012 ) Sequential changes in gene expression profiles in breast cancers during treatment with the aromatase inhibitor, letrozole. Pharmacogenom J 12(1):10–21
Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K et al (2012) PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res 14(1):1–9
Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D et al (2010) PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor–positive breast cancer. Proc Natl Acad Sci USA 107(22):10208–10213
Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK et al (2009) PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 15(16):5049–5059
Ellis MJ, Lin L, Crowder R, Tao Y, Hoog J, Snider J et al (2009) Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat 119(2):379
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M et al (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68(15):6084–6091
Papaxoinis G, Kotoula V, Alexopoulou Z, Kalogeras KT, Zagouri F, Timotheadou E et al (2015) Significance of PIK3CA mutations in patients with early breast cancer treated with adjuvant chemotherapy: a Hellenic Cooperative Oncology Group (HeCOG) study. PLoS ONE 10(10):e0140293
Chia YH, Ellis MJ, Ma CX (2010 Sep 7) Neoadjuvant endocrine therapy in primary breast cancer: indications and use as a research tool. Br J Cancer 103(6):759–764
Christiansen P, Ejlertsen B, Jensen M-B, Mouridsen H (2016) Danish breast cancer cooperative group. Clin Epidemiol 8:445
Skriver SK, Laenkholm A-V, Rasmussen BB, Handler J, Grundtmann B, Tvedskov TF et al (2018) Neoadjuvant letrozole for postmenopausal estrogen receptor-positive, HER2-negative breast cancer patients, a study from the Danish Breast Cancer Cooperative Group (DBCG). Acta Oncol 57(1):31–37
Skriver SK, Jensen M-B, Knoop AS, Ejlertsen B, Laenkholm A-V (2020) Tumour-infiltrating lymphocytes and response to neoadjuvant letrozole in patients with early oestrogen receptor-positive breast cancer: analysis from a nationwide phase II DBCG trial. Breast Cancer Res 22(1):46
Kaufmann M, von Minckwitz G, Mamounas EP, Cameron D, Carey LA, Cristofanilli M et al (2012) Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann Surg Oncol 19(5):1508–1516
Ogston KN, Miller ID, Payne S, Hutcheon AW, Sarkar TK, Smith I et al (2003) A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast 12(5):320–327
Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V et al (2007) Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 25(28):4414–4422
Bossuyt V, Provenzano E, Symmans WF, Boughey JC, Coles C, Curigliano G et al (2015) Recommendations for standardized pathological characterization of residual disease for neoadjuvant clinical trials of breast cancer by the BIG-NABCG collaboration. Ann Oncol 26(7):1280–1291
Residual Cancer Burden Calculator [Internet]. MD Anderson Cancer Center. https://www3.mdanderson.org/app/medcalc/index.cfm?pagename=jsconvert3. Accessed 7 Oct 2019
Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S et al (2010) American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. JCO 28(16):2784–2795
Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH et al (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013
Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J et al (2011) Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer Working Group. J Natl Cancer Inst 103(22):1656–1664
Dieci MV, Radosevic-Robin N, Fineberg S, van den Eynden G, Ternes N, Penault-Llorca F et al (2018) Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: a report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin Cancer Biol 1(52):16–25
Krupke DM, Begley DA, Sundberg JP, Richardson JE, Neuhauser SB, Bult CJ (2017) The Mouse Tumor Biology Database (MTB): a comprehensive resource for mouse models of human cancer. Cancer Res 77(21):e67–70
McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Online Mendelian Inheritance in Man, OMIM® [Internet]. https://omim.org/
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al (2018) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46(Database issue):D1062–D1067
Schemper M, Smith TL (1996) A note on quantifying follow-up in studies of failure time. Control Clin Trials 17(4):343–346
Loibl S, Volz C, Mau C, Blohmer J-U, Costa SD, Eidtmann H et al (2014) Response and prognosis after neoadjuvant chemotherapy in 1,051 patients with infiltrating lobular breast carcinoma. Breast Cancer Res Treat 144(1):153–162
Delpech Y, Coutant C, Hsu L, Barranger E, Iwamoto T, Barcenas CH et al (2013) Clinical benefit from neoadjuvant chemotherapy in oestrogen receptor-positive invasive ductal and lobular carcinomas. Br J Cancer 108(2):285–291
Ellsworth RE, Blackburn HL, Shriver CD, Soon-Shiong P, Ellsworth DL (2017) Molecular heterogeneity in breast cancer: state of the science and implications for patient care. Semin Cell Dev Biol 1(64):65–72
André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS et al (2019) Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 380(20):1929–1940
Verret B, Cortes J, Bachelot T, Andre F, Arnedos M (2019) Efficacy of PI3K inhibitors in advanced breast cancer. Ann Oncol 30(Suppl 10):x12–x20
Funding
This study was funded by The Danish Cancer Society (Grant No. R146-A9562), The Harboe Foundation, University of Copenhagen’s Foundation for cancer research (Grant No. 2019-0018) and The Sejer Persson and Lis Klüver foundation. The founding sources are all non-commercial and had no role in the design or execution of the study and have not reviewed the data or manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SKS declares she has no conflict of interest. MBJ has received institutional grants from Nanostring Technologies Inc and Oncology Venture. JOE declares he has no conflict of interest none, LBA declares she has no conflict of interest, ASK has received an institutional grant from Roche and is on advisory board for: Novartis, Astra Zeneca, MDS, Roche, Pfizer and ELI LILLY DANMARK A/S. MR is on advisory board for Astra Zeneca, BE has received institutional grants from: Nanostring Technologies Inc, Novartis and Roche. AVL has received research grants from Nanostring Technologies Inc. and Roche.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study is registered on clinicaltrials.gov (NCT00908531). The Biomedical Research Ethics of the Danish Capital Region approved the protocol (H-15012740). The genomic investigations presented here were subsequently approved (H-16031391).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Skriver, S.K., Jensen, MB., Eriksen, JO. et al. Induction of PIK3CA alterations during neoadjuvant letrozole may improve outcome in postmenopausal breast cancer patients. Breast Cancer Res Treat 184, 123–133 (2020). https://doi.org/10.1007/s10549-020-05833-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05833-w