Abstract
Background
While the prognostic relevance of lymphovascular invasion (LVI) in breast cancer is well known, its molecular biology is poorly understood. We hypothesized that pathologically determined LVI reflects molecular features of tumors and can be discerned from their genomic and transcriptomic profiles.
Methods
LVI status and Nottingham histological scores of primary breast tumors of The Cancer Genome Atlas (TCGA) project were assessed from pathology reports; other clinical and molecular data were obtained from TCGA data portals and publications. Two independent datasets (GSE5460 and GSE7849) were combined and used for validation.
Results
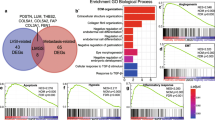
LVI status was determinable for 639 and 196 cases of the TCGA and validation cohorts, among whom LVI incidence was 37.8% and 37.2%, respectively. LVI was associated with high tumor Ki67 expression, advanced pathologic stage, and high Nottingham scores. LVI-positive cases had worse overall and progression-free survival regardless of cancer subtype. Surprisingly, in both cohorts, LVI was not associated with lymphangiogenesis or lymphatic vessel density as estimated from tumor expression of lymphatic endothelium-associated genes. LVI-positive tumors had higher genome copy number aberrations, aneuploidy, and homologous recombination defects, but not single-nucleotide variations or intra-tumor genome heterogeneity. Tumor immune cell composition and cytolytic activity was not associated with LVI status. On the other hand, expression of cell proliferation-related genes was significantly increased in LVI-positive tumors.
Conclusion
Our study suggests that breast cancer with LVI is a highly proliferative cancer, and it does not correlate with gene expression markers for lymphangiogenesis or immune response.






Similar content being viewed by others
Data availability
The GEO datasets used in this study are available at https://www.ncbi.nlm.nih.gov/geo with accession numbers GSE5460 and GSE7849. Clinical, gene-level mapped read counts of RNA sequencing data, and MAF files of TCGA-BRCA cases are available at Genome Data Commons portal of National Cancer Institute, USA at https://gdc.cancer.gov. Information on Nottingham scores and LVI status that was collated from pathology reports of the TCGA cases is in Table S2. Other data used in this study is available from sources cited in the Materials and Methods section.
Abbreviations
- CNA:
-
Copy number alteration
- ER:
-
Estrogen receptor
- GSEA:
-
Gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- HER2:
-
Human epidermal growth factor receptor 2
- HRD:
-
Homologous recombination defect
- LEC:
-
Lymphatic endothelial cell
- LVD:
-
Lymphatic vessel density
- LVI:
-
Lymphovascular invasion
- PR:
-
Progesterone receptor
- S1P:
-
Sphingosine-1-phosphate
- SNV:
-
Single-nucleotide variation
- TCGA:
-
The Cancer Genome Atlas
- TIES:
-
Text Information Extraction System
- TPM:
-
Transcripts per million
- tSNE:
-
T-distributed stochastic neighbor embedding
References
Ma Q, Dieterich LC, Detmar M (2018) Multiple roles of lymphatic vessels in tumor progression. Curr Opin Immunol 53:7–12. https://doi.org/10.1016/j.coi.2018.03.018
Zhang S, Zhang D, Gong M, Wen L, Liao C, Zou L (2017) High lymphatic vessel density and presence of lymphovascular invasion both predict poor prognosis in breast cancer. BMC Cancer 17(1):335. https://doi.org/10.1186/s12885-017-3338-x
Rakha EA, Martin S, Lee AH, Morgan D, Pharoah PD, Hodi Z, Macmillan D, Ellis IO (2012) The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer 118(15):3670–3680. https://doi.org/10.1002/cncr.26711
Zhang ZQ, Han YZ, Nian Q, Chen G, Cui SQ, Wang XY (2015) Tumor invasiveness, not lymphangiogenesis, is correlated with lymph node metastasis and unfavorable prognosis in young breast cancer patients (%3c/=35 years). PLoS ONE 10(12):e0144376. https://doi.org/10.1371/journal.pone.0144376
Zhang S, Zhang D, Yi S, Gong M, Lu C, Cai Y, Tang X, Zou L (2017) The relationship of lymphatic vessel density, lymphovascular invasion, and lymph node metastasis in breast cancer: a systematic review and meta-analysis. Oncotarget 8(2):2863–2873. https://doi.org/10.18632/oncotarget.13752
Enholm B, Karpanen T, Jeltsch M, Kubo H, Stenback F, Prevo R, Jackson DG, Yla-Herttuala S, Alitalo K (2001) Adenoviral expression of vascular endothelial growth factor-C induces lymphangiogenesis in the skin. Circ Res 88(6):623–629
Oh SJ, Jeltsch MM, Birkenhager R, McCarthy JE, Weich HA, Christ B, Alitalo K, Wilting J (1997) VEGF and VEGF-C: specific induction of angiogenesis and lymphangiogenesis in the differentiated avian chorioallantoic membrane. Dev Biol 188(1):96–109. https://doi.org/10.1006/dbio.1997.8639
Du Y, Liu H, He Y, Liu Y, Yang C, Zhou M, Wang W, Cui L, Hu J, Gao F (2013) The interaction between LYVE-1 with hyaluronan on the cell surface may play a role in the diversity of adhesion to cancer cells. PLoS ONE 8(5):e63463. https://doi.org/10.1371/journal.pone.0063463
Nagahashi M, Ramachandran S, Rashid OM, Takabe K (2010) Lymphangiogenesis: a new player in cancer progression. World J Gastroenterol 16(32):4003–4012. https://doi.org/10.3748/wjg.v16.i32.4003
Tammela T, Alitalo K (2010) Lymphangiogenesis: molecular mechanisms and future promise. Cell 140(4):460–476. https://doi.org/10.1016/j.cell.2010.01.045
Klahan S, Wong HS, Tu SH, Chou WH, Zhang YF, Ho TF, Liu CY, Yih SY, Lu HF, Chen SC, Huang CC, Chang WC (2017) Identification of genes and pathways related to lymphovascular invasion in breast cancer patients: a bioinformatics analysis of gene expression profiles. Tumour Biol 39(6):1010428317705573. https://doi.org/10.1177/1010428317705573
Paduch R (2016) The role of lymphangiogenesis and angiogenesis in tumor metastasis. Cell Oncol (Dordrecht) 39(5):397–410. https://doi.org/10.1007/s13402-016-0281-9
Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T (2018) The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer Sci 109(12):3671–3678. https://doi.org/10.1111/cas.13802
Tsuchida J, Nagahashi M, Nakajima M, Moro K, Tatsuda K, Ramanathan R, Takabe K, Wakai T (2016) Breast cancer sphingosine-1-phosphate is associated with phospho-sphingosine kinase 1 and lymphatic metastasis. J Surg Res 205(1):85–94. https://doi.org/10.1016/j.jss.2016.06.022
Jacobson RS, Becich MJ, Bollag RJ, Chavan G, Corrigan J, Dhir R, Feldman MD, Gaudioso C, Legowski E, Maihle NJ, Mitchell K, Murphy M, Sakthivel M, Tseytlin E, Weaver J (2015) A federated network for translational cancer research using clinical data and biospecimens. Cancer Res 75(24):5194–5201. https://doi.org/10.1158/0008-5472.Can-15-1973
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H, Cancer Genome Atlas Research N (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173(2):400–416. https://doi.org/10.1016/j.cell.2018.02.052
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404. https://doi.org/10.1158/2159-8290.CD-12-0095
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CE, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich L, Cancer Genome Atlas Research N (2018) The immune landscape of cancer. Immunity 48(4):812–830. https://doi.org/10.1016/j.immuni.2018.03.023
Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, Sofia HJ, Hutter C, Getz G, Wheeler D, Ding L, Group MCW, Cancer Genome Atlas Research N (2018) Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 6(3):271–281. https://doi.org/10.1016/j.cels.2018.03.002
Carlson J, Li JZ, Zollner S (2018) Helmsman: fast and efficient mutation signature analysis for massive sequencing datasets. BMC Genomics 19(1):845. https://doi.org/10.1186/s12864-018-5264-y
Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C (2016) DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17:31. https://doi.org/10.1186/s13059-016-0893-4
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259. https://doi.org/10.1007/978-1-4939-7493-1_12
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160(1–2):48–61. https://doi.org/10.1016/j.cell.2014.12.033
Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL (2008) Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat 108(2):191–201. https://doi.org/10.1007/s10549-007-9596-6
Anders CK, Acharya CR, Hsu DS, Broadwater G, Garman K, Foekens JA, Zhang Y, Wang Y, Marcom K, Marks JR, Mukherjee S, Nevins JR, Blackwell KL, Potti A (2008) Age-specific differences in oncogenic pathway deregulation seen in human breast tumors. PLoS ONE 3(1):e1373. https://doi.org/10.1371/journal.pone.0001373
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England) 28(6):882–883. https://doi.org/10.1093/bioinformatics/bts034
Podgrabinska S, Braun P, Velasco P, Kloos B, Pepper MS, Skobe M (2002) Molecular characterization of lymphatic endothelial cells. Proc Natl Acad Sci USA 99(25):16069–16074. https://doi.org/10.1073/pnas.242401399
Wick N, Saharinen P, Saharinen J, Gurnhofer E, Steiner CW, Raab I, Stokic D, Giovanoli P, Buchsbaum S, Burchard A, Thurner S, Alitalo K, Kerjaschki D (2007) Transcriptomal comparison of human dermal lymphatic endothelial cells ex vivo and in vitro. Physiol Genomics 28(2):179–192. https://doi.org/10.1152/physiolgenomics.00037.2006
Hirakawa S, Hong YK, Harvey N, Schacht V, Matsuda K, Libermann T, Detmar M (2003) Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am J Pathol 162(2):575–586. https://doi.org/10.1016/S0002-9440(10)63851-5
Amatschek S, Kriehuber E, Bauer W, Reininger B, Meraner P, Wolpl A, Schweifer N, Haslinger C, Stingl G, Maurer D (2007) Blood and lymphatic endothelial cell-specific differentiation programs are stringently controlled by the tissue environment. Blood 109(11):4777–4785. https://doi.org/10.1182/blood-2006-10-053280
DiMaio TA, Wentz BL, Lagunoff M (2016) Isolation and characterization of circulating lymphatic endothelial colony forming cells. Exp Cell Res 340(1):159–169. https://doi.org/10.1016/j.yexcr.2015.11.015
Petrova TV, Makinen T, Makela TP, Saarela J, Virtanen I, Ferrell RE, Finegold DN, Kerjaschki D, Yla-Herttuala S, Alitalo K (2002) Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. EMBO J 21(17):4593–4599
Aran D, Hu Z, Butte AJ (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18(1):220. https://doi.org/10.1186/s13059-017-1349-1
Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. https://doi.org/10.1186/1471-2105-14-7
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1(6):417–425. https://doi.org/10.1016/j.cels.2015.12.004
Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D'Eustachio P (2018) The reactome pathway knowledgebase. Nucleic Acids Res 46(D1):D649–D655. https://doi.org/10.1093/nar/gkx1132
Smyth G (2005) Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Dudoit S, Irizarry RA (eds) Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, pp 397–420
Ryu YJ, Kang SJ, Cho JS, Yoon JH, Park MH (2018) Lymphovascular invasion can be better than pathologic complete response to predict prognosis in breast cancer treated with neoadjuvant chemotherapy. Medicine 97(30):e11647. https://doi.org/10.1097/md.0000000000011647
Rakha EA, Abbas A, Pinto Ahumada P, ElSayed ME, Colman D, Pinder SE, Ellis IO (2018) Diagnostic concordance of reporting lymphovascular invasion in breast cancer. J Clin Pathol 71(9):802–805. https://doi.org/10.1136/jclinpath-2017-204981
Moynahan ME, Jasin M (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11(3):196–207. https://doi.org/10.1038/nrm2851
Lee AH, Pinder SE, Macmillan RD, Mitchell M, Ellis IO, Elston CW, Blamey RW (2006) Prognostic value of lymphovascular invasion in women with lymph node negative invasive breast carcinoma. Eur J Cancer (Oxford, England: 1990) 42(3):357–362. https://doi.org/10.1016/j.ejca.2005.10.021
Lauria R, Perrone F, Carlomagno C, De Laurentiis M, Morabito A, Gallo C, Varriale E, Pettinato G, Panico L, Petrella G et al (1995) The prognostic value of lymphatic and blood vessel invasion in operable breast cancer. Cancer 76(10):1772–1778
Schoppmann SF, Bayer G, Aumayr K, Taucher S, Geleff S, Rudas M, Kubista E, Hausmaninger H, Samonigg H, Gnant M, Jakesz R, Horvat R (2004) Prognostic value of lymphangiogenesis and lymphovascular invasion in invasive breast cancer. Ann Surg 240(2):306–312
Ejlertsen B, Jensen MB, Rank F, Rasmussen BB, Christiansen P, Kroman N, Kvistgaard ME, Overgaard M, Toftdahl DB, Mouridsen HT (2009) Population-based study of peritumoral lymphovascular invasion and outcome among patients with operable breast cancer. J Natl Cancer Inst 101(10):729–735. https://doi.org/10.1093/jnci/djp090
Bettelheim R, Penman HG, Thornton-Jones H, Neville AM (1984) Prognostic significance of peritumoral vascular invasion in breast cancer. Br J Cancer 50(6):771–777
Mohammed RA, Martin SG, Mahmmod AM, Macmillan RD, Green AR, Paish EC, Ellis IO (2011) Objective assessment of lymphatic and blood vascular invasion in lymph node-negative breast carcinoma: findings from a large case series with long-term follow-up. J Pathol 223(3):358–365. https://doi.org/10.1002/path.2810
Mohammed RA, Menon S, Martin SG, Green AR, Paish EC, Ellis IO (2014) Prognostic significance of lymphatic invasion in lymph node-positive breast carcinoma: findings from a large case series with long-term follow-up using immunohistochemical endothelial marker. Mod Pathol 27(12):1568–1577. https://doi.org/10.1038/modpathol.2014.60
Kurozumi S, Joseph C, Sonbul S, Alsaeed S, Kariri Y, Aljohani A, Raafat S, Alsaleem M, Ogden A, Johnston SJ, Aleskandarany MA, Fujii T, Shirabe K, Caldas C, Ashankyty I, Dalton L, Ellis IO, Desmedt C, Green AR, Mongan NP, Rakha EA (2019) A key genomic subtype associated with lymphovascular invasion in invasive breast cancer. Br J Cancer 120(12):1129–1136. https://doi.org/10.1038/s41416-019-0486-6
Cooper LA, Demicco EG, Saltz JH, Powell RT, Rao A, Lazar AJ (2018) PanCancer insights from The Cancer Genome Atlas: the pathologist's perspective. J Pathol 244(5):512–524. https://doi.org/10.1002/path.5028
Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Van Arnam J, Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V, Cancer Genome Atlas Research N (2018) Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep 23(1):181–193. https://doi.org/10.1016/j.celrep.2018.03.086
Shen S, Wu G, Xiao G, Du R, Hu N, Xia X, Zhou H (2018) Prediction model of lymphovascular invasion based on clinicopathological factors in Chinese patients with invasive breast cancer. Medicine 97(43):e12973. https://doi.org/10.1097/md.0000000000012973
Kanyilmaz G, Yavuz BB, Aktan M, Karaagac M, Uyar M, Findik S (2019) Prognostic Importance of Ki-67 in breast cancer and its relationship with other prognostic factors. Eur J Breast Health 15(4):256–261. https://doi.org/10.5152/ejbh.2019.4778
He KW, Sun JJ, Liu ZB, Zhuo PY, Ma QH, Liu ZY, Yu ZY (2017) Prognostic significance of lymphatic vessel invasion diagnosed by D2–40 in Chinese invasive breast cancers. Medicine 96(44):e8490. https://doi.org/10.1097/MD.0000000000008490
Feng Y, Wang W, Hu J, Ma J, Zhang Y, Zhang J (2010) Expression of VEGF-C and VEGF-D as significant markers for assessment of lymphangiogenesis and lymph node metastasis in non-small cell lung cancer. Anat Rec (Hoboken, NJ: 2007) 293(5):802–812. https://doi.org/10.1002/ar.21096
Ding M, Fu X, Tan H, Wang R, Chen Z, Ding S (2012) The effect of vascular endothelial growth factor C expression in tumor-associated macrophages on lymphangiogenesis and lymphatic metastasis in breast cancer. Mol Med Rep 6(5):1023–1029. https://doi.org/10.3892/mmr.2012.1043
Zhang S, Yi S, Zhang D, Gong M, Cai Y, Zou L (2017) Intratumoral and peritumoral lymphatic vessel density both correlate with lymph node metastasis in breast cancer. Sci Rep 7:40364. https://doi.org/10.1038/srep40364
Agarwal S, Singh A, Bagga PK (2018) Immunohistochemical evaluation of lymphovascular invasion in carcinoma breast with CD34 and D2–40 and its correlation with other prognostic markers. Indian J Pathol Microbiol 61(1):39–44. https://doi.org/10.4103/IJPM.IJPM_791_16
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434(7035):917–921. https://doi.org/10.1038/nature03445
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434(7035):913–917. https://doi.org/10.1038/nature03443
Rakha EA, Abd El Rehim D, Pinder SE, Lewis SA, Ellis IO (2005) E-cadherin expression in invasive non-lobular carcinoma of the breast and its prognostic significance. Histopathology 46(6):685–693. https://doi.org/10.1111/j.1365-2559.2005.02156.x
Corso G, Intra M, Trentin C, Veronesi P, Galimberti V (2016) CDH1 germline mutations and hereditary lobular breast cancer. Fam Cancer 15(2):215–219. https://doi.org/10.1007/s10689-016-9869-5
Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A, Zhao R, Milstien S, Zhou H, Spiegel S, Takabe K (2012) Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Can Res 72(3):726–735. https://doi.org/10.1158/0008-5472.can-11-2167
Miyan M, Schmidt-Mende J, Kiessling R, Poschke I, de Boniface J (2016) Differential tumor infiltration by T-cells characterizes intrinsic molecular subtypes in breast cancer. J Transl Med 14(1):227. https://doi.org/10.1186/s12967-016-0983-9
Eryilmaz MK, Mutlu H, Unal B, Salim DK, Musri FY, Coskun HS (2018) The importance of stromal and intratumoral tumor lymphocyte infiltration for pathologic complete response in patients with locally advanced breast cancer. J Cancer Res Ther 14(3):619–624. https://doi.org/10.4103/0973-1482.174550
Duijf PHG, Nanayakkara D, Nones K, Srihari S, Kalimutho M, Khanna KK (2019) Mechanisms of genomic instability in breast cancer. Trends Mol Med 25(7):595–611. https://doi.org/10.1016/j.molmed.2019.04.004
Mohammed RA, Martin SG, Gill MS, Green AR, Paish EC, Ellis IO (2007) Improved methods of detection of lymphovascular invasion demonstrate that it is the predominant method of vascular invasion in breast cancer and has important clinical consequences. Am J Surg Pathol 31(12):1825–1833. https://doi.org/10.1097/PAS.0b013e31806841f6
Su YL, Li SH, Chen YY, Chen HC, Tang Y, Huang CH, Chou FF, Wu SC, Rau KM (2014) Post-mastectomy radiotherapy benefits subgroups of breast cancer patients with T1–2 tumor and 1–3 axillary lymph node(s) metastasis. Radiol Oncol 48(3):314–322. https://doi.org/10.2478/raon-2013-0085
Funding
This study was supported by National Institutes of Health (NIH), USA grants R01CA160688 to K.T. and R25CA181003 to Roswell Park Comprehensive Cancer Center in support of F.Z.'s research internship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This study examined human data that had been generated in the past by other studies. Informed consent was therefore not obtained in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplementary material 1 (PDF 281 kb)
Text S1—Details of methods.Table S1—Characteristics of TCGA-BRCA tumors for which LVI status was determinable or not.Table S2—Nottingham histologic scores and LVI status for TCGA-BRCA tumors examined in this study.Table S3—Members of the endothelium-specific 88-gene and xCell gene sets.Table S4—Mutation frequency among LVI-positive and -negative tumors of TCGA-BRCA cohort for ten most commonly mutated genes.Table S5—mSigDb Hallmark and Reactome gene sets with significant enrichment in LVI-positive vs. -negative tumor comparison of TCGA-BRCA cohort by GSVA method.Table S6—mSigDb Hallmark and Reactome gene sets with significant enrichment in LVI-positive vs. -negative tumor comparison of validation cohort by GSVA method.Table S7—Top 20 each of genes whose expression is significantly down- or up-regulated in LVI-positive compared to -negative tumors of TCGA-BRCA cohort.
Rights and permissions
About this article
Cite this article
Asaoka, M., Patnaik, S.K., Zhang, F. et al. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res Treat 181, 309–322 (2020). https://doi.org/10.1007/s10549-020-05630-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05630-5