Abstract
Introduction
Large structured databases of pathology findings are valuable in deriving new clinical insights. However, they are labor intensive to create and generally require manual annotation. There has been some work in the bioinformatics community to support automating this work via machine learning in English. Our contribution is to provide an automated approach to construct such structured databases in Chinese, and to set the stage for extraction from other languages.
Methods
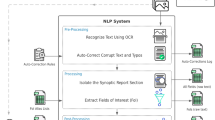
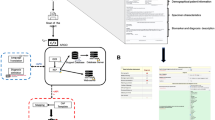
We collected 2104 de-identified Chinese benign and malignant breast pathology reports from Hunan Cancer Hospital. Physicians with native Chinese proficiency reviewed the reports and annotated a variety of binary and numerical pathologic entities. After excluding 78 cases with a bilateral lesion in the same report, 1216 cases were used as a training set for the algorithm, which was then refined by 405 development cases. The Natural language processing algorithm was tested by using the remaining 405 cases to evaluate the machine learning outcome. The model was used to extract 13 binary entities and 8 numerical entities.
Results
When compared to physicians with native Chinese proficiency, the model showed a per-entity accuracy from 91 to 100% for all common diagnoses on the test set. The overall accuracy of binary entities was 98% and of numerical entities was 95%. In a per-report evaluation for binary entities with more than 100 training cases, 85% of all the testing reports were completely correct and 11% had an error in 1 out of 22 entities.
Conclusion
We have demonstrated that Chinese breast pathology reports can be automatically parsed into structured data using standard machine learning approaches. The results of our study demonstrate that techniques effective in parsing English reports can be scaled to other languages.


Similar content being viewed by others
References
Huang CR, Chen KJ, Chang LL (1996) Segmentation standard for Chinese natural language processing. In: Proceedings of the 16th conference on Computational linguistics, vol. 2 (pp. 1045–1048). Association for Computational Linguistics
Wong KF, Li W, Xu R, Zhang ZS (2009) Introduction to Chinese natural language processing. Synth Lect Hum Lang Technol 2(1):1–148
Qiu X, Qi Z, Huang X (2013) Fudan NLP: a toolkit for Chinese natural language processing. In: ACL (conference system demonstrations), pp. 49–54
Liang YF, Chu PY, Chang CS, Wang CH, Chang P (2006) Developing and evaluating a simple, spreadsheet-based pathology report extraction system for cancer registrars. AMIA Ann Sym Proc 2006:1008
Buckley JM, Coopey SB, Sharko J, Polubriaginof F, Drohan B, Belli AK, Kim EM, Garber JE, Smith BL, Gadd MA et al (2012) The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform 3:23
Yala Adam, Barzilay Regina, Salama Laura, Griffin Molly, Sollender Grace, Bardia Aditya, Lehman Constance et al (2017) Using machine learning to parse breast pathology reports. Breast Cancer Res Treat 161(2):203–211
Sun J (2013) Jieba (version 0.39) [source code]. https://github.com/fxsjy/jieba
Korobov M (2015) Sklearn-crfsuite (Version 0.3.6) [source code] https://github.com/TeamHG-Memex/sklearn-crfsuite
Burger G, Abu-Hanna A, de Keizer N, Cornet R (2016) Natural language processing in pathology: a scoping review. J Clin Pathol 69(11):949–955
Edwards GA (2008) Expert systems for clinical pathology reporting. Clin Biochem Rev 29:S105–S109
Napolitano G, Fox C, Middleton R, Connolly D (2010) Pattern based information extraction from pathology reports for cancer registration. Cancer Causes Control 21:1887–1894
Nguyen A, Lawley M, Hansen D, Colquist S (2011) Structured pathology reporting for cancer from free text: lung cancer case study. Electron J Health Inform 7:8
Nguyen AN, Lawley MJ, Hansen DP, Bowman RV, Clarke BE, Duhig EE, Colquist S (2010) Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc 17:440–445
Weegar R, Dalianis H (2015) Creating a rule based system for text mining of Norwegian breast cancer pathology reports. In: Sixth international workshop on health text mining and information analysis (Louhi), p 73
Li Y, Martinez D (2010) Information extraction of multiple entities from pathology reports. In: Australasian Language Technology Association Workshop, p 41
Martinez D, Li Y (2011) Information extraction from pathology reports in a hospital setting. In: Proceedings of the 20th ACM international conference on information and knowledge management, ACM, pp 1877–1882
Nguyen A, Moore D, McCowan I, Courage M-J (2007) Multiclass classification of cancer stages from free-text histology reports using support vector machines. In: 29th annual international conference of the IEEE engineering in medicine and biology society, IEEE, pp 5140–5143
Wieneke AE, Bowles EJ, Cronkite D, Wernli KJ, Gao H, Carrell D, Buist DS (2015) Validation of natural language processing to extract breast cancer pathology procedures and results. J Pathol Inform 6:38
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tang, R., Ouyang, L., Li, C. et al. Machine learning to parse breast pathology reports in Chinese. Breast Cancer Res Treat 169, 243–250 (2018). https://doi.org/10.1007/s10549-018-4668-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-018-4668-3