Abstract
Purpose
We aimed to develop a highly sensitive method to detect ESR1 mutations in cell-free DNA (cfDNA) using next-generation sequencing with molecular barcode (MB–NGS) targeting the hotspot segment (c.1600–1713).
Methods
The sensitivity of MB–NGS was tested using serially diluted ESR1 mutant DNA and then cfDNA samples from 34 patients with metastatic breast cancer were analyzed with MB–NGS. The results of MB–NGS were validated in comparison with conventional NGS and droplet digital PCR (ddPCR).
Results
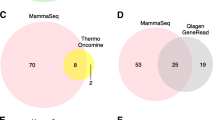
MB–NGS showed a higher sensitivity (0.1%) than NGS without barcode (1%) by reducing background errors. Of the cfDNA samples from 34 patients with metastatic breast cancer, NGS without barcode revealed seven mutations in six patients (17.6%) and MB–NGS revealed six additional mutations including three mutations not reported in the COSMIC database of breast cancer, resulting in total 13 ESR1 mutations in ten patients (29.4%). Regarding the three hotspot mutations, all the patients with mutations detected by MB–NGS had identical mutations detected by droplet digital PCR (ddPCR), and mutant allele frequency correlated very well between both (r = 0.850, p < 0.01). Moreover, all the patients without these mutations by MB–NGS were found to have no mutations by ddPCR.
Conclusion
In conclusion, MB–NGS could successfully detect ESR1 mutations in cfDNA with a higher sensitivity of 0.1% than conventional NGS and was considered as clinically useful as ddPCR.





Similar content being viewed by others
References
Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11(6):426–437. doi:10.1038/nrc3066
Diaz LA Jr, Bardelli A (2014) Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32(6):579–586. doi:10.1200/JCO.2012.45.2011
Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N (2013) Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497(7447):108–112. doi:10.1038/nature12065
Nakauchi C, Kagara N, Shimazu K, Shimomura A, Naoi Y, Shimoda M, Kim SJ, Noguchi S (2016) Detection of TP53/PIK3CA mutations in cell-free plasma dna from metastatic breast cancer patients using next generation sequencing. Clin Breast Cancer 16(5):418–423. doi:10.1016/j.clbc.2016.05.004
Katsnelson A (2013) Momentum grows to make ‘personalized’ medicine more ‘precise’. Nat Med 19(3):249. doi:10.1038/nm0313-249
Rothe F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, Fumagalli D, Michiels S, Drisis S, Moerman C, Detiffe JP, Larsimont D, Awada A, Piccart M, Sotiriou C, Ignatiadis M (2014) Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol 25(10):1959–1965. doi:10.1093/annonc/mdu288
Collin FS, Varmus H (2015) A new initiative on precision medicine. N Engl J Med 372(9):793–795
Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM (2008) Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med 14(5):579–584. doi:10.1038/nm1708
Mauger F, How-Kit A, Tost J (2017) COLD-PCR technologies in the area of personalized medicine: methodology and applications. Mol Diagn Ther. doi:10.1007/s40291-016-0254-8
Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW Jr, Alizadeh AA, Diehn M (2014) An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20(5):548–554. doi:10.1038/nm.3519
Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B (2011) Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA 108(23):9530–9535. doi:10.1073/pnas.1105422108
Kukita Y, Matoba R, Uchida J, Hamakawa T, Doki Y, Imamura F, Kato K (2015) High-fidelity target sequencing of individual molecules identified using barcode sequences: de novo detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. DNA Res 22(4):269–277. doi:10.1093/dnares/dsv010
Glenn TC (2011) Field guide to next-generation DNA sequencers. Mol Ecol Resour 11(5):759–769. doi:10.1111/j.1755-0998.2011.03024.x
Villaflor V, Won B, Nagy R, Banks K, Lanman RB, Talasaz A, Salgia R (2016) Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 7(41):66880–66891. doi:10.18632/oncotarget.11801
Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, Gursky A, Siddiqui J, Tomlins SA, Roychowdhury S, Pienta KJ, Kim SY, Roberts JS, Rae JM, Van Poznak CH, Hayes DF, Chugh R, Kunju LP, Talpaz M, Schott AF, Chinnaiyan AM (2013) Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45(12):1446–1451. doi:10.1038/ng.2823
Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, Hudis C, Chen D, Taran T, Hortobagyi G, Greene G, Berger M, Baselga J, Chandarlapaty S (2013) ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45(12):1439–1445. doi:10.1038/ng.2822
Segal CV, Dowsett M (2014) Estrogen receptor mutations in breast cancer–new focus on an old target. Clin Cancer Res 20(7):1724–1726. doi:10.1158/1078-0432.CCR-14-0067
Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, Bhatt T, Patel P, Voi M, Gnant M, Hortobagyi G, Baselga J, Moynahan ME (2016) Prevalence of ESR1 mutations in cell-free dna and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol 2(10):1310–1315. doi:10.1001/jamaoncol.2016.1279
Fribbens C, O’Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, Cristofanilli M, Andre F, Loi S, Loibl S, Jiang J, Bartlett CH, Koehler M, Dowsett M, Bliss JM, Johnston SR, Turner NC (2016) Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol 34(25):2961–2968. doi:10.1200/JCO.2016.67.3061
Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, Aimi J, Derynck MK, Chen M, Chan IT, Amler LC, Hampton GM, Johnston S, Krop I, Schmid P, Lackner MR (2016) Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 7:11579. doi:10.1038/ncomms11579
Niu J, Andres G, Kramer K, Kundranda MN, Alvarez RH, Klimant E, Parikh AR, Tan B, Staren ED, Markman M (2015) Incidence and clinical significance of ESR1 mutations in heavily pretreated metastatic breast cancer patients. Onco Targets Ther 8:3323–3328. doi:10.2147/OTT.S92443
Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL, De Stanchina E, Carlson KE, Martin TS, Uddin S, Li Z, Fanning S, Katzenellenbogen JA, Greene G, Baselga J, Chandarlapaty S (2017) Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov 7(3):277–287. doi:10.1158/2159-8290.CD-15-1523
Yanagawa T, Kagara N, Miyake T, Tanei T, Naoi Y, Shimoda M, Shimazu K, Kim SJ, Noguchi S (2017) Detection of ESR1 mutations in plasma and tumors from metastatic breast cancer patients using next-generation sequencing. Breast Cancer Res Treat. doi:10.1007/s10549-017-4190-z
Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, Smith A, Leux C, Garcia-Murillas I, Ferraldeschi R, Lorente D, Mateo J, Ong M, Yap TA, Banerji U, Gasi Tandefelt D, Turner N, Attard G, de Bono JS (2015) Serial next-generation sequencing of circulating cell-free dna evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 21(20):4586–4596. doi:10.1158/1078-0432.CCR-15-0584
Perkins G, Yap TA, Pope L, Cassidy AM, Dukes JP, Riisnaes R, Massard C, Cassier PA, Miranda S, Clark J, Denholm KA, Thway K, Gonzalez De Castro D, Attard G, Molife LR, Kaye SB, Banerji U, de Bono JS (2012) Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS ONE 7(11):e47020. doi:10.1371/journal.pone.0047020
Takai E, Totoki Y, Nakamura H, Morizane C, Nara S, Hama N, Suzuki M, Furukawa E, Kato M, Hayashi H, Kohno T, Ueno H, Shimada K, Okusaka T, Nakagama H, Shibata T, Yachida S (2015) Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep 5:18425. doi:10.1038/srep18425
Allred DC, Harvey JM, Berardo M, Clark GM (1998) Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 11(2):155–168
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30(5):614–620. doi:10.1093/bioinformatics/btt593
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. doi:10.1093/bioinformatics/bts565
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. doi:10.1038/nmeth.1923
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. doi:10.1093/bioinformatics/btr509
Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, Hadfield J, May AP, Caldas C, Brenton JD, Rosenfeld N (2012) Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 4(136):136ra68. doi:10.1126/scitranslmed.3003726
Peng Q, Vijaya Satya R, Lewis M, Randad P, Wang Y (2015) Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics 16:589. doi:10.1186/s12864-015-1806-8
Stahlberg A, Krzyzanowski PM, Jackson JB, Egyud M, Stein L, Godfrey TE (2016) Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res 44(11):e105. doi:10.1093/nar/gkw224
Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I, Lopez-Knowles E, Ribas R, Nerurkar A, Osin P, Chandarlapaty S, Martin LA, Dowsett M, Smith IE, Turner NC (2015) Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 7(313):313ra182. doi:10.1126/scitranslmed.aac7551
Acknowledgements
The authors would like to thank Katsuhide Yoshidome (Osaka Police Hospital, Osaka, Japan) for kindly providing the plasma samples of healthy volunteers examined in this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
This study was supported in part by the research funding from Novartis, Pfizer, and Sysmex. Shinzaburo Noguchi has received research grant for other study from AstraZeneca and honoraria from AstraZeneca, Novartis, Pfizer, and Sysmex, and has been an advisor for AstraZeneca and Novartis. Naofumi Kagara received honoraria from AstraZeneca and Novartis. Yasuto Naoi received honoraria from AstraZeneca and Sysmex and research grant from AstraZeneca for other study. Masafumi Shimoda received honoraria from Novartis. Kenzo Shimazu received honoraria from AstraZeneca. Seung Jin Kim received honoraria from AstraZeneca, Novartis, and Pfizer. The other authors declare that they do not have a financial relationship with the organizations that sponsored the research.
Ethical approval
This study complied with the current laws of Japan. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2017_4487_MOESM2_ESM.pptx
Supplementary Fig. 1 The clinical course of the patients with ESR1 mutations. The clinical course and the treatments are shown in the panel for the ten patients with ESR1 mutations. Treatments with aromatase inhibitors are highlighted in orange. LN, lymph node; met, metastasis; Ax, axillary; Sc; supraclavicular; abd. Abdominal; FEC, fluorouracil, epirubicin and cyclophosphamide; PTX, paclitaxel; LET, letrozole; TAM, tamoxifen; RT, radiation therapy; TOR, toremifene; ANA, anastrozole; EXE, exemestane; Bev, bevacizumab; ERI, eribulin; MPA, medroxyprogesterone acetate; DOC, docetaxel; GnRHa, gonadotropin releasing hormone analog; Zola (Zoladex), goserelin acetate; GEM, gemcitabine; VNR, vinorelbine; CPT-11, camptothecin 11; NK105, paclitaxel-incorporating micellar nanoparticle formulation; FUL, fulvestrant; Leu (Leuplin), leuprorelin; CMF, cyclophosphamide, methotrexate and fluorouracil; CE, cyclophosphamide and epirubicin; Lap, lapatinib; CAP, capecitabine; EVE, everolimus; XC, capecitabine and cyclophosphamide; FUR, doxifluridine; Tzb, trastuzumab; DOC + T+P, docetaxel, trastuzumab and pertuzumab (PPTX 50 Skb)
Rights and permissions
About this article
Cite this article
Masunaga, N., Kagara, N., Motooka, D. et al. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res Treat 167, 49–58 (2018). https://doi.org/10.1007/s10549-017-4487-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4487-y