Abstract
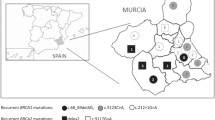
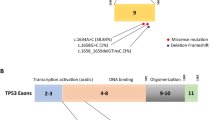
In this study, we analyzed a “variant of uncertain significance” (VUS) located in exon 23 of the BRCA2 gene exhibited by six members of five distinct families with hereditary breast cancer (BC). The variant was identified by DNA sequencing, and cDNA analysis revealed its co-expression with wild-type mRNA. We analyzed co-occurrence with other pathological mutations in BRCA1/2, performed a case–control study, looked for evolutionary data and used in-silico analyses to predict its potential clinical significance. Sequencing revealed an in frame deletion of 126 nucleotides in exon 23, leading to a deletion of 42 amino acids (c.9203_9328del126, p.Pro2992_Thr3033del). All of the VUS-carriers suffered from either BC or ovarian/pancreatic cancer. No other definite pathologic mutation of BRCA genes was found in the five families. The identified deletion could not be observed in a control cohort of 2,652 healthy individuals, but in 5 out of 916 (0.5%) tested BC families without a bona fide pathogenic BRCA1/2 mutation (P = 0.0011). According to these results, the in frame deletion c.9203_9328del126 is a rare mutation strongly associated with familial BC. In summary, our investigations indicate that this BRCA2 deletion is pathogenic.



Similar content being viewed by others
Abbreviations
- VUS:
-
Variant of uncertain significance
- UCV or UV:
-
Unclassified variant
- BRCA:
-
Breast cancer gene
- BC:
-
Breast cancer
- OC:
-
Ovarian cancer
- PC:
-
Pancreatic cancer
- LC:
-
Lung cancer
- NMD:
-
Nonsense-mediated RNA decay
References
Abkevich V, Zharkikh A, Deffenbaugh AM, Frank D, Chen Y, Shattuck D, Skolnick MH, Gutin A, Tavtigian SV (2004) Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet 41:492–507
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72:1117–1130. doi:10.1086/375033
Arnold N, Peper H, Bandick K, Kreikemeier M, Karow D, Teegen B, Jonat W (2002) Establishing a control population to screen for the occurrence of nineteen unclassified variants in the BRCA1 gene by denaturing high-performance liquid chromatography. J Chromatogr B 782:99–104
Billack B, Monteiro AN (2004) Methods to classify BRCA1 variants of uncertain clinical significance: the more the merrier. Cancer Biol Ther 3:458–459
Biswas K, Das R, Alter BP, Kuznetsov SG, Stauffer S, North SL, Burkett S, Brody LC, Meyer S, Byrd RA, Sharan SK (2011) A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood 118:2430–2442. doi:10.1182/blood-2010-12-324541
Borg A, Haile RW, Malone KE, Capanu M, Diep A, Torngren T, Teraoka S, Begg CB, Thomas DC, Concannon P, Mellemkjaer L, Bernstein L, Tellhed L, Xue S, Olson ER, Liang X, Dolle J, Borresen-Dale AL, Bernstein JL (2010) Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE study. Hum Mutat 31:E1200–E1240. doi:10.1002/humu.21202
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913–917. doi:10.1038/nature03443
Campos B, Diez O, Domenech M, Baena M, Balmana J, Sanz J, Ramirez A, Alonso C, Baiget M (2003) RNA analysis of eight BRCA1 and BRCA2 unclassified variants identified in breast/ovarian cancer families from Spain. Hum Mutat 22:337. doi:10.1002/humu.9176
Chenevix-Trench G, Healey S, Lakhani S, Waring P, Cummings M, Brinkworth R, Deffenbaugh AM, Burbidge LA, Pruss D, Judkins T, Scholl T, Bekessy A, Marsh A, Lovelock P, Wong M, Tesoriero A, Renard H, Southey M, Hopper JL, Yannoukakos K, Brown M, Easton D, Tavtigian SV, Goldgar D, Spurdle AB (2006) Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res 66:2019–2027. doi:10.1158/0008-5472.CAN-05-3546
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162:156–159. doi:10.1006/abio.1987.9999
Consortium BCL (1999) Cancer risks in BRCA2 mutation carriers. The Breast Cancer Linkage Consortium. J Natl Cancer Inst 91:1310–1316
Cote S, Arcand SL, Royer R, Nolet S, Mes-Masson AM, Ghadirian P, Foulkes WD, Tischkowitz M, Narod SA, Provencher D, Tonin PN (2011) The BRCA2 c.9004G>A (E2003K) variant is likely pathogenic and recurs in breast and/or ovarian cancer families of French Canadian descent. Breast Cancer Res Treat. doi:10.1007/s10549-011-1796-4
Coyne RS, McDonald HB, Edgemon K, Brody LC (2004) Functional characterization of BRCA1 sequence variants using a yeast small colony phenotype assay. Cancer Biol Ther 3:453–457
Deffenbaugh AM, Frank TS, Hoffman M, Cannon-Albright L, Neuhausen SL (2002) Characterization of common BRCA1 and BRCA2 variants. Genet Test 6:119–121. doi:10.1089/10906570260199375
Domchek S, Weber BL (2008) Genetic variants of uncertain significance: flies in the ointment. J Clin Oncol 26:16–17. doi:10.1200/JCO.2007.14.4154
Figge MA, Blankenship L (2004) Missense mutations in the BRCT domain of BRCA-1 from high-risk women frequently perturb strongly hydrophobic amino acids conserved among mammals. Cancer Epidemiol Biomarkers Prev 13:1037–1041
Finch A, Beiner M, Lubinski J, Lynch HT, Moller P, Rosen B, Murphy J, Ghadirian P, Friedman E, Foulkes WD, Kim-Sing C, Wagner T, Tung N, Couch F, Stoppa-Lyonnet D, Ainsworth P, Daly M, Pasini B, Gershoni-Baruch R, Eng C, Olopade OI, McLennan J, Karlan B, Weitzel J, Sun P, Narod SA (2006) Salpingo-oophorectomy and the risk of ovarian, fallopian tube, and peritoneal cancers in women with a BRCA1 or BRCA2 mutation. JAMA 296:185–192. doi:10.1001/jama.296.2.185
Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA (2003) Understanding missense mutations in the BRCA1 gene: an evolutionary approach. Proc Natl Acad Sci USA 100:1151–1156. doi:10.1073/pnas.0237285100
Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Zelada-Hedman M et al (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet 62:676–689
Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, Campra D, Grutzmann R, Rehder H, Rothmund M, Schmiegel W, Neoptolemos JP, Bartsch DK (2003) BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst 95:214–221
Howard HJ, Horaitis O, Cotton RG, Vihinen M, Dalgleish R, Robinson P, Brookes AJ, Axton M, Hoffmann R, Tuffery-Giraud S (2010) The Human Variome Project (HVP) 2009 forum “towards establishing standards”. Hum Mutat 31:366–367. doi:10.1002/humu.21175
Karchin R, Monteiro AN, Tavtigian SV, Carvalho MA, Sali A (2007) Functional impact of missense variants in BRCA1 predicted by supervised learning. PLoS Comput Biol 3:e26. doi:10.1371/journal.pcbi.0030026
Kuznetsov SG, Liu P, Sharan SK (2008) Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med 14:875–881. doi:10.1038/nm.1719
Machackova E, Foretova L, Lukesova M, Vasickova P, Navratilova M, Coene I, Pavlu H, Kosinova V, Kuklova J, Claes K (2008) Spectrum and characterisation of BRCA1 and BRCA2 deleterious mutations in high-risk Czech patients with breast and/or ovarian cancer. BMC Cancer 8:140. doi:10.1186/1471-2407-8-140
Malacrida S, Agata S, Callegaro M, Casella C, Barana D, Scaini MC, Manoukian S, Oliani C, Radice P, Barile M, Menin C, D’Andrea E, Montagna M (2008) BRCA1 p.Val1688del is a deleterious mutation that recurs in breast and ovarian cancer families from Northeast Italy. J Clin Oncol 26:26–31. doi:10.1200/JCO.2007.13.2118
Marroni F, Aretini P, D’Andrea E, Caligo MA, Cortesi L, Viel A, Ricevuto E, Montagna M, Cipollini G, Federico M, Santarosa M, Marchetti P, Bailey-Wilson JE, Bevilacqua G, Parmigiani G, Presciuttini S (2004) Penetrances of breast and ovarian cancer in a large series of families tested for BRCA1/2 mutations. Eur J Hum Genet 12:899–906. doi:10.1038/sj.ejhg.5201256
Marroni F, Aretini P, D’Andrea E, Caligo MA, Cortesi L, Viel A, Ricevuto E, Montagna M, Cipollini G, Ferrari S, Santarosa M, Bisegna R, Bailey-Wilson JE, Bevilacqua G, Parmigiani G, Presciuttini S (2004) Evaluation of widely used models for predicting BRCA1 and BRCA2 mutations. J Med Genet 41:278–285
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Mirkovic N, Marti-Renom MA, Weber BL, Sali A, Monteiro AN (2004) Structure-based assessment of missense mutations in human BRCA1: implications for breast and ovarian cancer predisposition. Cancer Res 64:3790–3797. doi:10.1158/0008-5472.CAN-03-3009
Narod S (2002) What options for treatment of hereditary breast cancer? Lancet 359:1451–1452. doi:10.1016/S0140-6736(02)08465-9
Narod SA, Boyd J (2002) Current understanding of the epidemiology and clinical implications of BRCA1 and BRCA2 mutations for ovarian cancer. Curr Opin Obstet Gynecol 14:19–26
Osorio A, de la Hoya M, Rodriguez-Lopez R, Martinez-Ramirez A, Cazorla A, Granizo JJ, Esteller M, Rivas C, Caldes T, Benitez J (2002) Loss of heterozygosity analysis at the BRCA loci in tumor samples from patients with familial breast cancer. Int J Cancer 99:305–309. doi:10.1002/ijc.10337
Ostrow KL, McGuire V, Whittemore AS, DiCioccio RA (2004) The effects of BRCA1 missense variants V1804D and M1628T on transcriptional activity. Cancer Genet Cytogenet 153:177–180. doi:10.1016/j.cancergencyto.2004.01.020
Peelen T, van Vliet M, Bosch A, Bignell G, Vasen HF, Klijn JG, Meijers-Heijboer H, Stratton M, van Ommen GJ, Cornelisse CJ, Devilee P (2000) Screening for BRCA2 mutations in 81 Dutch breast-ovarian cancer families. Br J Cancer 82:151–156. doi:10.1054/bjoc.1999.0892
Radice P, De Summa S, Caleca L, Tommasi S (2011) Unclassified variants in BRCA genes: guidelines for interpretation. Ann Oncol 22(Suppl 1):i18–i23. doi:10.1093/annonc/mdq661
Ramirez CJ, Fleming MA, Potter JD, Ostrander GK, Ostrander EA (2004) Marsupial BRCA1: conserved regions in mammals and the potential effect of missense changes. Oncogene 23:1780–1788. doi:10.1038/sj.onc.1207292
Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, Jack E, Vesprini DJ, Kuperstein G, Abrahamson JL, Fan I, Wong B, Narod SA (2001) Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet 68:700–710. doi:10.1086/318787
Schmutzler RK, Rhiem K, Breuer P, Wardelmann E, Lehnert M, Coburger S, Wappenschmidt B (2006) Outcome of a structured surveillance programme in women with a familial predisposition for breast cancer. Eur J Cancer Prev 15:483–489. doi:10.1097/01.cej.0000220624.70234.14
Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, Timmerman MM, Brody LC, Tucker MA (1997) The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med 336:1401–1408. doi:10.1056/NEJM199705153362001
Szabo C, Masiello A, Ryan JF, Brody LC (2000) The breast cancer information core: database design, structure, and scope. Hum Mutat 16:123–131. doi:10.1002/1098-1004(200008)16:2<123:AID-HUMU4>3.0.CO;2-Y
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A (2006) Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43:295–305. doi:10.1136/jmg.2005.033878
Thompson D, Easton DF, Goldgar DE (2003) A full-likelihood method for the evaluation of causality of sequence variants from family data. Am J Hum Genet 73:652–655. doi:10.1086/378100
Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M, Carmichael J (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376:235–244. doi:10.1016/S0140-6736(10)60892-6
Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondum-Nielsen K, Gerdes AM, Moller P, Kristoffersson U, Olsson H, Borg A, Monteiro AN (2001) Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet 10:353–360
Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295:1379–1388. doi:10.1001/jama.295.12.1379
Williams RS, Chasman DI, Hau DD, Hui B, Lau AY, Glover JN (2003) Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J Biol Chem 278:53007–53016. doi:10.1074/jbc.M310182200
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378:789–792. doi:10.1038/378789a0
Wu K, Hinson SR, Ohashi A, Farrugia D, Wendt P, Tavtigian SV, Deffenbaugh A, Goldgar D, Couch FJ (2005) Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res 65:417–426
Yang Y, Swaminathan S, Martin BK, Sharan SK (2003) Aberrant splicing induced by missense mutations in BRCA1: clues from a humanized mouse model. Hum Mol Genet 12:2121–2131. doi:10.1093/hmg/ddg222
Acknowledgments
We thank the families and the blood donors for providing samples for this study. We also acknowledge the support of Dr. Rongxi Yang and the excellent technical assistance of Claudia Dinkelacker, Yvonne Kilian, Kerstin Lang, Gloria Luta, and Sabine Serick. This study was supported by the Dietmar-Hopp Foundation, the Helmholtz-Society, the German Cancer Aid, and the German Cancer Research Center (DKFZ).
Conflict of interest
All authors declared that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Michelle G. Rath, Farnoosh Fathali-Zadeh, Christian Sutter and Barbara Burwinkel contributed equally to this work.
Rights and permissions
About this article
Cite this article
Rath, M.G., Fathali-Zadeh, F., Langheinz, A. et al. Molecular and clinical characterization of an in frame deletion of uncertain clinical significance in the BRCA2 gene. Breast Cancer Res Treat 133, 725–734 (2012). https://doi.org/10.1007/s10549-011-1917-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1917-0