Abstract
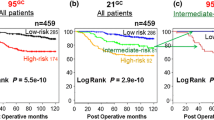
Our aim was to develop an accurate diagnostic system using gene expression analysis by means of DNA microarray for prognosis of node-negative and estrogen receptor (ER)-positive breast cancer patients in order to identify a subset of patients who can be safely spared adjuvant chemotherapy. A diagnostic system comprising a 95-gene classifier was developed for predicting the prognosis of node-negative and ER-positive breast cancer patients by using already published DNA microarray (gene expression) data (n = 549) as the training set and the DNA microarray data (n = 105) obtained at our institute as the validation set. Performance of the 95-gene classifier was compared with that of conventional prognostic factors as well as of the genomic grade index (GGI) based on the expression of 70 genes. With the 95-gene classifier we could classify the 105 patients in the validation set into a high-risk (n = 44) and a low-risk (n = 61) group with 10-year recurrence-free survival rates of 93 and 53%, respectively (P = 8.6e−7). Multivariate analysis demonstrated that the 95-gene classifier was the most important and significant predictor of recurrence (P = 9.6e−4) independently of tumor size, histological grade, progesterone receptor, HER2, Ki67, or GGI. The 95-gene classifier developed by us can predict the prognosis of node-negative and ER-positive breast cancer patients with high accuracy. The 95-gene classifier seems to perform better than the GGI. As many as 58% of the patients classified into the low-risk group with this classifier could be safely spared adjuvant chemotherapy.


Similar content being viewed by others
Abbreviations
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
References
van’t Veer LJ, Dai H, Van de Vijver MJ et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536
Paik S (2007) Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. Oncologist 12:631–635
Sotiriou C, Wirapati P, Loi S et al (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98:262–272
Morimoto K, Kim SJ, Tanei T et al (2009) Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci 100:1062–1068
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19:403–410
Yixin W, Profjan GM, Yi Z et al (2005) Gene-expression profile to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365:634–635
Lance DM, Johanna S, Joshy G et al (2005) An expression signature for p53 status in human breast cancer predicts mutant status, transcriptional effects, and patient survival. PNAS 102:13550–13555
Sherene L, Christine D, Benjamin HK et al (2007) Definition of clinically distant molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 25:1239–1246
Christine D, Fanny P, Sherene L et al (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicancer independent validation series. Clin Cancer Res 13:3207–3214
Sherene L, Benjamin HK, Christine D et al (2008) Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 9:239
Rafael AI, Bridget H, Francois C et al (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Choi JK, Yu U, Kim S et al (2003) Combining multiple microarray studies and modeling interstudy variation. Bioinformatics 19(Suppl 1):i84–i90
Desmedt C, Giobbie-Hurder A, Neven P et al (2009) The Gene expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1–98 trial. BMC Med Genomics 2:40
Haibe-Kains B, Desmedt C, Piette F et al (2008) Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics 9:394
Stec J, Wang J, Coombes K et al (2005) Comparison of the predictive accuracy of DNA array-based multigene classifiers across cDNA arrays and Affymetrix GeneChips. J Mol Diagn 7:357–367
Bueno-de-Mesquita JM, Linn SC, Keijzer R et al (2009) Validation of 70-gene prognosis signature in node-negative breast cancer. Breast Cancer Res Treat 117:483–495
Ishitobi M, Goranova TE, Komoike Y et al (2010) Clinical utility of the 70-gene MammaPrint profile in a Japanese population. Jpn J Clin Oncol 40:508–512
Buyse M, Loi S, van’t Veer L et al (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 98:1183–1192
Wittner BS, Sgroi DC, Ryan PD et al (2008) Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res 14:2988–2993
Sparano JA, Paik S (2008) Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol 26:721–728
Mamounas EP, Tang G, Fisher B et al (2010) Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol 28:1677–1683
Albain KS, Paik S, van’t Veer L (2009) Prediction of adjuvant chemotherapy benefit in endocrine responsive, early breast cancer using multigene assays. Breast 18(Suppl 3):S141–S145
Sparano JA (2006) TAILORx: trial assigning individualized options for treatment (Rx). Clin Breast Cancer 7:347–350
Zujewski JA, Kamin L (2008) Trial assessing individualized options for treatment for breast cancer: the TAILORx trial. Future Oncol 4:603–610
Acknowledgments
The authors acknowledge Knowledge Cluster Initiative, Scientific Research on Priority Areas programs of the Ministry of Education, Culture, Sports, Science and Technology of Japan and Comprehensive 10-Year Strategy for Cancer Control program of the Ministry of Health, Labour and Welfare, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naoi, Y., Kishi, K., Tanei, T. et al. Development of 95-gene classifier as a powerful predictor of recurrences in node-negative and ER-positive breast cancer patients. Breast Cancer Res Treat 128, 633–641 (2011). https://doi.org/10.1007/s10549-010-1145-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-1145-z