Abstract
Systems biology has provided new resources for discovering and reasoning about mechanisms. In addition to generating databases of large bodies of data, systems biologists have introduced platforms such as Cytoscape to represent protein–protein interactions, gene interactions, and other data in networks. Networks are inherently flat structures. One can identify clusters of highly connected nodes, but network representations do not represent these clusters as at a higher level than their constituents. Mechanisms, however, are hierarchically organized: they can be decomposed into their parts and their activities can be decomposed into component operations. A potent bridge between flat networks and hierarchical mechanisms is provided by biological ontologies, both those curated by hand such as Gene Ontology (GO) and those extracted directly from databases such as Network Extracted Ontology (NeXO). I examine several examples in which by applying ontologies to networks, systems biologists generate new hypotheses about mechanisms and characterize these novel strategies for developing mechanistic explanations.

From Constanzo et al. (2010), Figure 1, reprinted with permission from AAAS. (Color figure online)

From Constanzo et al. (2010), Figure 2, reprinted with permission from AAAS. (Color figure online)

Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics from Ashburner et al. (2000). (Color figure online)

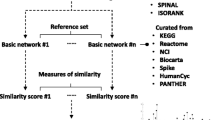
Reprinted by permission from Macmillan Publishers Ltd: Nature Biotechnology, from Dutkowski et al. (2013), Figure 1. (Color figure online)

Reprinted by permission from Macmillan Publishers Ltd: Nature Biotechnology, from Dutkowski et al. (2013), Figure 2. (Color figure online)

Reprinted by permission from Macmillan Publishers Ltd: Nature Biotechnology, from Dutkowski et al. (2013), Figure 4b. (Color figure online)

Reprinted from Kramer et al. (2017), Figure 2, with permission from Elsevier. (Color figure online)

From Kramer (2016), Chapter 4, Figure 6. (Color figure online)

Reprinted from Kramer et al. (2017), Figure 7, with permission from Elsevier. (Color figure online)

Reprinted from Yu et al. (2016), Figure 1, with permission from Elsevier. (Color figure online)

Reprinted from Yu et al. (2016), Figure 4, with permission from Elsevier. (Color figure online)
Similar content being viewed by others
Notes
Over the past 15 years numerous large databases based on a variety of interactions between genes or between molecules in cells have been made publically available. A regularly updated compilation of molecular biology databases is maintained at https://www.oxfordjournals.org/our_journals/nar/database/c/. It currently includes 1685 databases. Starting with supplementary issues in April 1991 and May 1992 and a regular issue in July, 1993, the journal Nucleic Acids Research has regularly reviewed databases. Beginning in 1996, the journal identified its first issue of each year as the database issue.
MGI was itself the product of integrating two mouse databases. In 2000 the Arabidopsis Information Resource (TAIR) and the Caenorhabditis elegans group joined GO.
“These particular classifications were chosen because they represent information sets that are common to all living forms and are basic to our annotation of information about genes and gene products” (Ashburner et al. 2001).
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Ashburner M, Ball CA, Blake JA, Butler H, Cherry JM, Corradi J, Dolinski K, Eppig JT, Harris M, Hill DP, Lewis S, Marshall B, Mungall C, Reiser L, Rhee S, Richardson JE, Richter J, Ringwald M, Rubin GM, Sherlock G, Yoon J, Consortium GO (2001) Creating the gene ontology resource: design and implementation. Genome Res 11:1425–1433
Bechtel W, Abrahamsen A (2005) Explanation: a mechanist alternative. Stud Hist Philos Biol Biomed Sci 36:421–441
Bechtel, W., & Richardson, R. C. (1993/2010). Discovering complexity: Decomposition and localization as strategies in scientific research. Cambridge, MA: MIT Press. 1993 edition published by Princeton University Press
Braillard P-A (2010) Systems biology and the mechanistic framework. Hist Philos Life Sci 32:43–62
Chervitz SA, Hester ET, Ball CA, Dolinski K, Dwight SS, Harris MA, Juvik G, Malekian A, Roberts S, Roe T, Scafe C, Schroeder M, Sherlock G, Weng S, Zhu Y, Cherry JM, Botstein D (1999) Using the saccharomyces genome database (SGD) for analysis of protein similarities and structure. Nucleic Acids Res 27:74–78
Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C (2010) The genetic landscape of a cell. Science 327:425–431
Craver CF, Darden L (2013) In search of mechanisms: discoveries across the life sciences. University of Chicago Press, Chicago
Dutkowski J, Kramer M, Surma MA, Balakrishnan R, Cherry JM, Krogan NJ, Ideker T (2013) A gene ontology inferred from molecular networks. Nat Biotechnol 31:38–45
Gene Ontology Consortium (2015) Gene Ontology Consortium: going forward. Nucleic Acids Res 43:D1049–D1056
Gross F (2011) What systems biology can tell us about disease. Hist Philos Life Sci 33:477–496
Gruber TR (1995) Toward principles for the design of ontologies used for knowledge sharing. Int J Hum Comput Stud 43:907–928
Guénolé A, Srivas R, Vreeken K, Wang ZZ, Wang SY, Krogan NJ, Ideker T, van Attikum H (2013) Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol Cell 49:346–358
Huang S (2011) Systems biology of stem cells: three useful perspectives to help overcome the paradigm of linear pathways. Philos Trans R Soc Lond B Biol Sci 366:2247–2259
Huneman P (2010) Topological explanations and robustness in biological sciences. Synthese 177:213–245
Ideker T, Krogan Nevan J (2012) Differential network biology. Mol Syst Biol 8:565
Kramer MH (2016) Transformation of high-throughput data into hierarchical cellular models enables biological prediction and discovery. Ph.D. Biomedical Science, University of California, San Diego, LaJolla, CA
Kramer MH, Dutkowski J, Yu MK, Bafna V, Ideker T (2014) Inferring gene ontologies from pairwise similarity data. Bioinformatics 30:i34–42
Kramer MH, Farre J-C, Mitra K, Yu MK, Ono K, Demchak B, Licon K, Flagg M, Balakrishnan R, Cherry JM, Subramani S, Ideker T (2017) Active interaction mapping reveals the hierarchical organization of autophagy. Mole Cell 65(761–774):e765
Lee I, Li Z, Marcotte EM (2007) An improved, bias-reduced probabilistic functional gene network of baker’s yeast, Saccharomyces cerevisiae. PLoS ONE 2:e988
Leonelli S (2010) Documenting the emergence of bio-ontologies: or, why researching bioinformatics requires HPSSB. Hist Philos Life Sci 32:105–125
Leonelli S (2012) Classificatory theory in data-intensive science: the case of open biomedical ontologies. Int Stud Phil Sci 26:47–65
Leonelli S, Diehl AD, Christie KR, Harris MA, Lomax J (2011) How the gene ontology evolves. BMC Bioinf 12:325
Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431:308–312
Machamer P, Darden L, Craver CF (2000) Thinking about mechanisms. Philos Sci 67:1–25
Park Y, Bader JS (2011) Resolving the structure of interactomes with hierarchical agglomerative clustering. BMC Bioinf 12:S44
Rubin GM, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LSB, Gong FC, Guan P, Harris NL, Hay BA, Hoskins RA, Li JY, Li ZY, Hynes RO, Jones SJM, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O’Farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XQH, Zhong F, Zhong WY, Gibbs R, Venter JC, Adams MD, Lewis S (2000) Comparative genomics of the eukaryotes. Science 287:2204–2215
Yu Michael K, Kramer M, Dutkowski J, Srivas R, Licon K, Kreisberg Jason F, Ng Cherie T, Krogan N, Sharan R, Ideker T (2016) Translation of genotype to phenotype by a hierarchy of cell subsystems. Cell Syst 2:77–88
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bechtel, W. Using the hierarchy of biological ontologies to identify mechanisms in flat networks. Biol Philos 32, 627–649 (2017). https://doi.org/10.1007/s10539-017-9579-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10539-017-9579-x