Abstract
The pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection involves dysregulations of iron metabolism, and although the mechanism of this pathology is not yet fully understood, correction of iron metabolism pathways seems a promising pharmacological target. The previously observed effect of inhibiting SARS-CoV-2 infection by ferristatin II, an inducer of transferrin receptor 1 (TfR1) degradation, prompted the study of competition between Spike protein and TfR1 ligands, especially lactoferrin (Lf) and transferrin (Tf). We hypothesized molecular mimicry of Spike protein as cross-reactivity of Spike-specific antibodies with Tf and Lf. Thus, strong positive correlations (R2 > 0.95) were found between the level of Spike-specific IgG antibodies present in serum samples of COVID-19-recovered and Sputnik V-vaccinated individuals and their Tf-binding activity assayed with peroxidase-labeled anti-Tf. In addition, we observed cross-reactivity of Lf-specific murine monoclonal antibody (mAb) towards the SARS-CoV-2 Spike protein. On the other hand, the interaction of mAbs produced to the receptor-binding domain (RBD) of the Spike protein with recombinant RBD protein was disrupted by Tf, Lf, soluble TfR1, anti-TfR1 aptamer, as well as by peptides RGD and GHAIYPRH. Furthermore, direct interaction of RBD protein with Lf, but not Tf, was observed, with affinity of binding estimated by KD to be 23 nM and 16 nM for apo-Lf and holo-Lf, respectively. Treatment of Vero E6 cells with apo-Lf and holo-Lf (1–4 mg/mL) significantly inhibited SARS-CoV-2 replication of both Wuhan and Delta lineages. Protective effects of Lf on different arms of SARS-CoV-2-induced pathogenesis and possible consequences of cross-reactivity of Spike-specific antibodies are discussed.
Similar content being viewed by others
Introduction
Pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) started in late 2019 and so far claimed the lives of more than 6 million people. Today coronavirus disease 2019 (COVID-19) remains a serious public health problem (Aleem et al. 2022). Poor understanding of molecular mechanisms of infection and its complications, as well as the lack of remedies against SARS-CoV-2 with proven efficacy requires that research on this virus be continued. The studies on the design of SARS-CoV-2 antivirals are complicated by the presence of multiple receptors on a host cell surface that the virus uses to enter the cell, such as angiotensin-converting enzyme 2 (ACE2), neuropilin-1, CD147, tyrosine-protein kinase receptor UFO (AXL) and other co-receptors (Scialo et al. 2020; Shang et al. 2020; Zhang et al. 2020; Wang et al. 2020a, 2021; Wei et al. 2020; Daly et al. 2020; Cantuti-Castelvetri et al. 2020; Jackson et al. 2022). Dysregulation of iron metabolism in SARS-CoV-2-infected patients, which is associated with onset of hypoxia, inflammation, and the response to oxidative stress has been widely reviewed (Cavezzi et al. 2022; Naidu et al. 2022; Kronstein-Wiedemann et al. 2022; Suriawinata and Mehta 2022). In this regard, the therapeutic potential of lactoferrin (Lf), a cationic homologue of serum transferrin (Tf), has been widely discussed as a regulator of inflammation, iron metabolism, tolerance to hypoxia and oxidative stress. Suriawinata and Mehta (2022) first discussed SARS-CoV-2-related dysregulations of iron metabolism in view of the transferrin receptor (TfR1) involvement in the infection (Tang et al. 2020a, b). Interestingly, the severity of COVID-19 is shadowed by the level of ferritin (Suriawinata and Mehta 2022), the heavy-chain form of which also interacts with TfR1. However, the binding site for ferritin is different from that for Tf (Sakamoto et al. 2015).
Interaction of TfR1 and ACE2 with Spike protein, excessive susceptibility to SARS-CoV-2 of mice transgenic for human TfR1, and protective effects of antibodies against TfR1 that reduce the severity of pathological changes in the lungs of infected mice have been demonstrated (Tang et al. 2020a, b). We pursued this area of research by testing the effect of TfR1 degradation on infection of Vero cells by SARS-CoV-2. Thus, we observed dose-dependent inhibition of SARS-CoV-2 replication (IC50 ~ 6–40 μM), as well as abrogated endocytosis of the receptor-binding domain (RBD) of Spike protein after treatment of cell culture with ferristatin II (Sokolov et al. 2022a, b). This compound induced degradation of TfR1 (Byrne et al. 2013), and was initially discovered in screening small-size inhibitors of iron influx into cancer cells (Brown et al. 2004). Byrne and colleagues (2013) hypothesized that ferristatin II releases human hemochromatosis protein (HFE) from its complex with TfR1 (Lebron et al. 1999), which results in TfR1 degradation and up-regulation of hepcidin. Although ferristatin II (also known as direct black 38, Chlorazol black E, C.I. 30,235) was non-toxic for cultured Vero cells and effectively altered iron metabolism in mice and rats, it is degraded to benzidine metabolites and cannot be used directly in humans (Dewan et al. 1988; Römer et al. 2014; Alkhateeb et al. 2015). Nevertheless, inhibiting SARS-CoV-2 entry by targeting TfR1 on the host cells, rather than the viral mutation-prone Spike protein, seems to be a promising therapeutic strategy.
The key role of TfR1 in cancer cell growth and TfR1-mediated penetration through the blood–brain barrier prompted the studies of several “anti-cancer” and “brain-directed” TfR1-binding molecules and their derivates, including Tf and Lf conjugates, synthetic peptides, i.e. HAIYPRH (also known as T7), aptamers, and monoclonal antibodies (mAbs) (Luck and Mason 2013; Mahidhara et al. 2015; Liang et al. 2018; Ayo et al. 2021; Candelaria et al. 2021; Zhang et al. 2022). RGD motif (integrin-binding motif) of TfR1 is involved in binding of Tf and Lf, as was demonstrated in structural studies of the complex formed by Tf and TfR1 (Dubljevic et al. 1999; Xu et al. 2005; Sakamoto et al. 2006). A mutation (R646H/G647A) in RGD-motif of TfR1 reduced cellular uptake of holo-Tf, but had no effect on heavy-chain ferritin uptake (Sakamoto et al. 2015). Noteworthy, the RBD of Spike protein of all SARS-CoV-2 lineages also contains RGD motif, whereas it is absent in previous human coronavirus variants (Dakal 2021; Makowski et al. 2021; Othman et al. 2022; Norris et al. 2022). Moreover, the interaction of Spike protein with bovine Lf has been reported in at least three studies (Campione et al. 2021; Miotto et al. 2021; Cutone et al. 2022). Several studies demonstrated protective effects of Lf (recombinant and milk-derived) against SARS-CoV-2 infection in vitro (Campione et al. 2021; Prieto-Fernández et al. 2021; Salaris et al. 2021; Mirabelli et al. 2021; Hu et al. 2021; Lai et al. 2022; Worting et al. 2022; Cutone et al. 2022). Different mechanisms of antiviral effect of Lf have been proposed, but not all papers contain information on the degree of iron saturation of Lf. In neutrophils granules, milk and other exocrine secretions, Lf is present predominantly in the apo-form (Masson et al. 1966; Britigan et al. 1994). The affinity of Lf to Fe(III) is much less affected by a decrease in pH, as compared with Tf, which is usually attributed to the noticeable bacteriostatic effect of apo-Lf (Brock 1980; Baker and Baker 2004). In the last two decades the anti-anaemic effect of Lf and its involvement in iron metabolism, especially in regulation of IL-6 and hepcidin effects, have been extensively studied (Cutone et al. 2014; Rosa et al. 2017; Lepanto et al. 2019; Artym et al. 2021). Iron-saturation of Lf affects the expression of TfR1 gene, i.e. apo-form of Lf up-regulates TfR1 via hypoxia-inducible factor (HIF) pathway; in contrast, holo-Lf down-regulates the expression of TfR1 (Zhang et al. 2021). It is worth noting that Tf and Lf also interact with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) which mediates Tf uptake and a rapid response to hypoxia (Kumar et al. 2012; Rawat et al. 2012; Malhotra et al. 2019). Other recent reviews on Lf structure and functions include its interactions with intelectin-1 (omentin-1), CD14, chemokine receptor 4 (CXCR4), and low-density lipoprotein receptor-related protein (LRP) in cellular receptors list, but interaction of Lf with TfR1 received little attention (Li and Guo 2021; Artym et al. 2021; Suzuki et al. 2005; Lepanto et al. 2019; Elzoghby et al. 2020; Godínez-Chaparro et al. 2021) or was fully neglected (Kawakawi et al. 1990; Mahidhara et al. 2015; Kell et al. 2020; Bartolomé et al. 2022). Interestingly, TfR1 is regarded as cellular receptor of Lf in a recent survey of its antiviral activity, and the corresponding scheme is presented (Ward et al. 2022). However, participation of TfR1 in interaction with SARS-CoV-2 is not mentioned, although the paper largely deals with an impact of viral infections on iron metabolism and describes the mechanisms of antiviral effect of Lf against SARS-CoV-2, in particular of the protein from cow milk (Ward et al. 2022). Taking into account the homology of Tf and Lf and their interaction with RGD-motif of TfR1, our study was focused on Tf and Lf, despite that the list of natural ligands of TfR1 also includes HFE, heavy-chain ferritin, haem-albumin (Jennifer et al. 2020) etc. Considering TfR1-HFE-mediated up-regulation of hepcidin and high levels of hepcidin and ferritin in serum samples obtained from severe COVID-19-infected patients (Zhou et al. 2020), we cannot exclude that COVID-19 infection alters the interaction of TfR1 with HFE and/or heavy-chain ferritin.
We hypothesized a molecular mimicry of SARS-CoV-2 Spike protein with Tf and Lf. To test this hypothesis, we examined the cross-reactivity of antibodies against Spike protein, Tf and Lf; the competition between Spike protein and synthetic TfR1 ligands, as well as Tf and Lf; and finally, the anti-viral effects of apo- and holo-Lf.
Materials and methods
Materials
The following reagents were used in the study: anti-TfR polyclonal rabbit antibody (ab84036, Abcam, UK); bovine serum albumin (BSA), dimethyl sulfoxide (DMSO), horseradish peroxidase (HRP) with RZ > 3.0 (Amresco, USA); recombinant SARS-CoV-2 Spike protein (1-1208 a.a., trimer, AtaGenix Laboratories, Wuhan, China); Bio-Gel A-1.5 m, Chelex-100, skimmed milk protein (blotting grade), HRP-labeled antibody against mouse IgG, HRP-labeled antibody against human IgG, HRP-labeled antibody against rabbit IgG, Tween 20, UNOsphere Q, UNOsphere S, (Bio-Rad, USA); acrylamide, ammonium persulfate, glycerol, glycine, mercaptoethanol, N,N′-methylene-bis-acrylamide, N,N,N′N′-tetramethylethylene diamine, Tris, NaCl, KCl, (NH4)2SO4, H3BO3 (Helicon, Russia); HCl and H2SO4 (Lenreactiv, Russia); N-hydroxysuccinimide ester of 5-TAMRA (Lumiprobe, Russia); biotinamidohexanoic acid N-hydroxysuccinimide, biotin hydrazide, CM-Sepharose, Coomassie R-250, phenylmethylsulfonyl fluoride SDS, Sephadex G-75 Superfine, soybean trypsin inhibitor (STI), 3,3′,5,5′-tetramethylbenzidine, Triton X-100, trypsin from bovine pancreas (Sigma Aldrich, USA), DEAE-Toyopearl 650-M (Toyosoda, Japan).
A recombinant protein corresponding to the receptor-binding domain (RBD) of SARS-CoV-2 S protein (Prokofyev et al. 2021) was expressed in eucaryotic cells by BIOCAD JSC (St. Petersburg, Russia). Aptamer against TfR1 Cy5-ACTCATAGGGTTAGGGGCTGCTGGCCAGATACTCAGATGGTAGGGTTACTATGAGC (Zhang et al. 2022) was synthesized by the company “Evrogen” (Russia). Peptides RGD and GHAIYPRH were synthesized by solid-phase synthesis on a 2-chlorotrityl chloride resin according to the Fmoc/tBu strategy using the carbodiimide method starting from the C-terminus. The peptides were purified from side products by RP-HPLC. To confirm the expected molecular mass the products obtained were analyzed by mass spectrometry.
Solutions for chromatography and surface plasmon resonance assay were prepared using apyrogenic deionized water with specific resistance 18.2 MΩ × cm. Following buffers were used: 150 mM NaCl, 10 mM Na-borate buffer, pH 8.3—BBS; 150 mM NaCl, 10 mM Na-phosphate buffer, pH 7.4—PBS; BBS and PBS, supplemented by 0,05% Tween 20—BBS-T, PBS-T; 150 mM NaCl, 10 mM Hepes–NaOH, pH 7.4 (HBS).
Viruses
Two strains of SARS-CoV-2 were obtained from the repository of Smorodintsev Research Institute of Influenza (Saint-Petersburg, Russia): hCoV-19/St_Petersburg-3524S/2020 (lineage B.1, Wuhan) and hCoV-19/Russia/SPE-RII-32759 V/2021 (lineage B.1.617.2, Delta). Virus stocks were generated by infecting Vero-E6 cells at a multiplicity of infection (MOI) 0.01 using DMEM supplemented with 2% FBS, 10 mM HEPES and 1 × antibiotic–antimycotic (all from Capricorn, Ebsdorfer-grund, Germany). After incubation for 72 h, cell supernatants were harvested, subjected to low-speed centrifugation to remove cell debris and stored in single-use aliquots at − 70 °C. Viral titers were determined by infecting 96-well plates seeded with Vero E6 cells, as described in (Amanat et al. 2020), and expressed in log10TCID50/mL.
Isolation and purification of proteins
Recombinant human apo-Lf purified from the milk of transgenic goats was obtained at the Belarusian State University and RUE «Scientific and Practical Centre for Animal Production of the National Academy of Sciences of Belarus». The product is officially branded “CAPRABEL™” and contains about 90% of the iron-free form of Lf (Semak et al. 2019). Tf was isolated from human blood plasma by ion-exchange chromatography on DEAE-Toyopearl, UNOsphere S and gel filtration on Sephadex G-75 Superfine (Sokolov et al. 2017). Apo-form of Tf was obtained by adding 2 mM ascorbic acid to 8 mg/mL Tf in 100 mM Na-acetate buffer, pH 5.5, the protein was immediately dialyzed against buffer containing 100 mM Na2HPO4 and 50 mM citric acid (pH 4.8), followed by the buffer replacement with HBS using centrifuge concentrators (Vivaspin 20, cut off 30 kDa). Holo-forms of Lf and Tf were obtained by adding 0.8 mM Fe(III) (10 mM stock solution of NH4Fe(SO4)2·12H2O in 100 mM Na-acetate buffer, pH 5.5) to 8 mg/mL of proteins in 200 mM Na-phosphate-citrate buffer (pH 8.0), followed by dialysis against the same buffer and filtration on a column with Chelex-100 equilibrated with HBS.
To obtain an affinity sorbent for TfR1 purification we used holo-Tf immobilized on Sepharose 6B activated by BrCN (about 5 mg holo-Tf per 1 mL of wet gel). TfR1 was purified from human placenta with small modifications of earlier protocols (Seligman and Allen 1987; Turkewitz et al. 1988; Kanevsky et al. 1997)—see Supplementary data, section S1. Soluble form of TfR (sTfR) was obtained by limited proteolysis of TfR with trypsin and by ion-exchange chromatography—see Supplementary data, section S2. Identification of TfR and sTfR by MALDI-TOF-tryptic fingerprint was performed—see Supplementary data, section S3, Figure S1.
Obtaining rabbit polyclonal antibodies against Tf and murine mAbs against human Lf and SARS-CoV-2 spike protein
To obtain polyclonal antibodies against Tf, rabbits were immunized with 300 μg of purified human holo-Tf, which was additionally cleared from admixtures by disc-electrophoresis in detergent-free polyacrylamide gel (see “Electrophoresisandwestern blotting”) with single-well comb for sample loading. After electrophoresis the narrow red band corresponding to holo-Tf was cut out of gel. Half of the gel was homogenized with equal volume of PBS and emulsion obtained was injected intracutaneously with 1/10 volume of Freund’s adjuvant (complete and incomplete for the first and the second injection, respectively). The second half of the gel was frozen awaiting the second injection. On the 8th, 12th, and 16th day after the second injection blood was collected, and separated serum was used for isolating IgG by ammonium sulphate fractionation and affinity chromatography on Tf-Sepharose. Purified antibodies were tested for specific reaction with human Tf in samples of human serum and plasma (Western-blotting and ELISA Enzyme-linked immunosorbent assay (ELISA)).
For obtaining mAbs, the hybridoma technology was used (Pandey et al. 2010). Briefly, 20 μg of Lf or Spike protein with complete Freund’s adjuvant per mouse (BALB/c) were used for the first immunization, and after 1 month 5 μg for the second. Then, on the 4th day, abdominal lymph nodes were removed and lymphocytes were mixed with SP2/0-AG14 cells (2:1). After hybridization with polyethylene glycol (1500) cells were seeded in 96-well cultural plates with one-day peritoneal macrophages. Hybrids were selected in RMPI-1640 medium supplemented with 10% fetal bovine serum and HAT-reagent. Enzyme-linked immunosorbent assay (ELISA) with Lf or Spike protein, and BSA as negative control were used for primary selection of clones (“ELISA”). Selected clones (4 for Lf and 6 for Spike protein) were subcloned, and individual clones were cultivated for injecting into mice (2 million cells per animal) and obtaining ascites. After 2 weeks ascitic fluids collected from the mice were used for purification of antibodies. Saturation by ammonium sulphate and affinity chromatography on a resin with immobilized Protein A allowed to obtain no less than 96% pure IgGs according to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis (see “Electrophoresisandwestern blotting”). mAbs against Lf were labeled according to the code of a well (3E11 and 3H5), while mAbs against Spike protein were designated as S1–S6. Selection of mAbs reacting with RBD of Spike protein was performed by ELISA (“ELISA”).
Labeling of proteins
Spike protein was labeled by linking biotin to primary amino groups (Spike-A-Bi) using biotinamidohexanoic acid N-hydroxysuccinimide ester and to carbohydrate moiety (Spike-C-Bi) using sodium periodate oxidation of carbohydrates and biotin hydrazide. Labeling of proteins (IgG, Lf anf Tf) with HRP was performed by sodium periodate oxidation of HRP carbohydrates and conjugation with proteins, followed by reduction of Shiff’s bases with NaBH4. Obtained HRP-labeled proteins were stored at − 20 °C in 50% glycerol.
Labeling by 5-TAMRA was carried out at + 4 °C for 12 h by adding 5 mM solution of N-hydroxysuccinimide ester of 5-TAMRA in DMSO to 30 µM apo-Lf, apo-Tf, holo-Lf, holo-Tf, or RBD solutions in 200 mM NaHCO3, keeping the ratio 5-TAMRA:protein = 8:1 (mol/mol). After removal of excessive dye, the degree of labeling was estimated by the ratio of A541 to A280, which ranged from 3.2 to 3.8 mol TAMRA per mol protein.
Recruiting participants and collecting blood samples
55 serum samples were collected from 53 COVID-19-recovered patients aged 21 to 84 years who participated in the study of SARS-CoV-2-specific humoral and T-cell responses (Matyushenko et al. 2021). The study was approved by the Ethics Committee of the Institute of Experimental Medicine (protocol No 2/20 dated 7 April 2020). Serum was sampled 1–12 months past recovery. A cohort of 24 COVID-19 naïve participants (aged 24 to 91 years) was used as a control group. A cohort of 16 Sputnik V-vaccinated individuals (aged 27–74 years) was used as a positive control group. Serum was sampled 1–4 months past vaccination. Serum samples were collected from individuals to assay anti-Spike and anti-RBD IgG, and Tf-binding capacity of these IgGs (“ELISA”). IgGs from representative samples of each cohort were purified by Protein A affinity chromatography for further Tf- and Lf-binding analyzes.
Analytical methods
Spectrophotometry
Optical spectra were registered on SF-2000-02 spectrophotometer («OKB-Spectr», Russia). Concentrations of purified apo-Lf, apo-Tf, and Fe(III)-saturated holo-forms were measured using the following extinction coefficients: apo-form Lf—ε280 = 87,360 M−1 cm−1, apo-form Tf—ε280 = 89,155 M−1 cm−1, holo-form Lf—ε465 = 4267 M−1 cm−1, holo-form Tf—ε465 = 4446 M−1 cm−1 (Masson 1970).
ELISA
Specific conditions of different variants of direct and competitive ELISA performed are summarized in Table 1. In all cases, polystyrene 96-well plates with high-binding capacity were incubated with 100 μL of immobilized protein (antigen or primary antibody against analyte detected in the assay) in BBS at 4 °C in a humidified chamber. Other reagents added into the wells were incubated in a thermoshaker (BioSan, 350 rpm, 37 °C, commonly for 1 h). Prior to the addition of the next reagent, the plates were washed with BBS-T or PBS-T in an automatic plate washer (PW-40, Bio-Rad). To prevent non-specific binding, 100 μL of a blocking reagent were added: 3% BSA or 3% skimmed milk protein in BBS-T or PBS-T (incubation for 1 h). Then, the analyte-containing samples (interacting with immobilized protein alone or with a competitor) were added. Next, either a chromogenic mixture (in case of competitive assay of HRP-labeled analyte) or HRP-labeled streptavidin (in case of biotin-labeled analyte) or HRP-labeled specific antibody was added. For HRP-labeled reagents, 100 μL of a chromogenic mixture containing 0.5 mM 3,3′,5,5′-tetramethylbenzidine and 2 mM hydrogen peroxide in 50 mM Na-acetate buffer (pH 5.5) were added. After development of the blue color (5–30 min), the reaction was terminated by adding 50 μL of 1 M H2SO4. Absorbance at 450 nm was registered in a microplate reader CLARIOstar (BMG Labtech, Germany).
ELISA of mouse IgG against Lf and spike protein in culture medium for selection of hybridomas
For selection of hybridomas (“Obtaining rabbit polyclonal antibodies against Tf and murine mAbs against human Lf and SARS-CoV-2 spike protein”), secretion of IgGs against Lf or Spike protein in culture medium was directly assayed. The level of mAbs against coated Lf or Spike protein was measured using HRP-labeled anti-mouse IgG (Table 1). BSA was also used as a blocking agent and coating protein (without antigens) for negative selection of hybridomas. mAbs raised against Spike protein and also capable of reacting with RBD were selected in a direct assay with HRP-labeled anti-mouse IgG of their binding with RBD or Spike protein on solid phase.
ELISA of IgGs against spike protein and RBD, and of binding human Tf with IgGs in serum samples
Each sample of serum was used in triplicate in four types of assays. Spike protein or RBD was immobilized on duplicate plates, followed by incubation with serum samples diluted 1:25. One of these plates was stained for IgGs (HRP-labeled human anti-IgG), while the other plate was stained for reaction of IgG with Tf (HRP-labeled anti-Tf) (see Table 1). The repeats of ELISA might vary due to numerous factors, starting with the time interval before the reaction is stopped and the fluctuations of the immobilized protein, and ending with the degree of dilution of samples and stock solutions of HRP-labeled reagents. A450 was repeatedly measured in 93 samples corresponding to the number of patients under study. For the control of intra-assay variations, each series of measurements included two duplicate samples obtained from patients recovered from COVID-19 and a duplicate sample from naïve donor. Those samples served as the highest, the medium, and the lowest point in the distribution of data. Upon normalization of the data obtained, these three duplicate samples were excluded from the overall results. The final IgG antibody level was defined as the A450 value multiplied by serum dilution (× 25).
ELISA of human Tf
Human Tf was assayed in sandwich ELISA with polyclonal rabbit anti-Tf on solid phase (Table 1). HRP-labeled polyclonal antibodies were applied after incubation of tested samples in serial dilutions and of Tf standards (1–4000 ng/mL).
ELISA of binding human Lf with IgGs and Tf obtained from serum samples
Binding of human Lf was assayed in sandwich ELISA with HRP-labeled Lf and IgGs or Tf on solid phase (Table 1). HRP-labeled Lf was applied after immobilization of IgGs or purified Tf on solid phase (5 mg/mL).
ELISA of spike protein binding with Lf and mAbs against Lf
Binding of Spike protein with Lf and mAbs against Lf was studied in a competitive assay. Two different mAbs against Lf (3H5 or 3E11) were immobilized on solid phase. Aliquots of Spike protein labeled by biotin to primary amino groups (Spike-A-Bi) or to carbohydrate moiety (Spike-C-Bi) were added to Lf in different concentrations (Table 1). Cooperative binding of biotin-labeled Spike protein with Lf and mAbs against Lf, or its competition with increasing Lf concentrations for binding to mAbs against Lf was detected using HRP-streptavidin conjugate.
ELISA of competitors of anti-spike mAb binding with RBD
Different ligands of TfR and sTfR were studied as competitors of interaction between mAb against Spike (S6) with RBD immobilized on solid phase. Binding of HRP-labeled mAb against Spike with different concentrations of sTfR (0.7–44 nM), Tf or Lf (10–640 nM), aptamer against TfR1 (50–500 nM), peptides RGD (2–120 μM) and GHAIYPRH (0.33–21 μM) was tested (Table 1). Results of analysis were presented as the dependence of binding on the concentration of a competitor.
Electrophoresis and western blotting
Proteins and their complexes were resolved under non-denaturing conditions by detergents-free disc-electrophoresis in polyacrylamide gel (disc-PAGE) (David 1964). Molecular weight of proteins and their homogeneity were analyzed by electrophoresis in SDS-containing gel in high-molar Tris buffer (SDS-PAGE) (Fling and Gregerson 1986). Proteins were identified by Western blotting (Anderson et al. 1982). After semi-dry transfer, the nitrocellulose membrane with proteins was kept for 30 min in 3% solution of dried skimmed milk in PBS-T. The membrane was left for 1 h or overnight for incubation with a primary antibody, followed by 2-h incubation with HRP-labeled secondary antibodies (1:1000). Bands that bound secondary antibodies were revealed after incubation of the membrane in the chromogenic solution obtained by mixing 2 ml of methanol containing 6 mg of 4-chloro-1-naphtol with 10 ml of 5 mM H2O2 in PBS.
Surface plasmon resonance assay binding of Lf to RBD
SPR measurements were carried out at 25 °C using a carboxymethylated-dextran chip (CM5) on dual flowcell Biacore × 100 instrument (GE-Healthcare, Piscataway, NJ, USA). The sensor chip was first activated with an equimolar (0.2 M) mixture of N-ethyl-N-dimethylaminopropylcarbodiimide and N-hydroxysuccinimide and then reacted with a solution of RBD (230 μg/mL) in 10 mM sodium acetate buffer, pH 5.5, injected at a flow rate of 5 μl/min for 12 min. Unreacted carboxymethylic groups on the sensor chip were blocked by 1 M ethanolamine at pH 8.5. Final immobilization levels of 3913 resonance units (RU) were obtained, corresponding to approximately 3.9 ng of bound RBD/mm2. Thereafter, increasing concentrations of apo-Lf and holo-Lf (4–256 nM) were injected over the RBD-coated sensor chip at a flow rate of 10 μl/min in 150 mM NaCl, 10 mM Hepes–NaOH, pH 7.5, 0.05% polyoxyethylene sorbitan (HBS-P +). Each run consisted of four steps: (1) buffer injection for 1 min; (2) Lf injection for 1 min; (3) dissociation for 5 min; (4) regeneration for 90 s with HBS-P + , containing 1 M NaCl. The reference flow cell, with no RBD immobilized, was used as a control to evaluate non-specific binding (< 2% of Rmax) of Lf to the sensor chip. The resulting control curves were subtracted from the corresponding binding curves recorded at each Lf concentration. The equilibrium dissociation constant, KD, of RBD-Lf complex was determined by plotting the steady-state value of RU as a function of Lf concentration, and the data were fitted to the Langmuir equation describing the one-site binding model. Kinetic parameters (kon—association rate constant, koff—dissociation rate constant, KD—equilibrium dissociation constant, and Rmax—A binding capacity of the surface) were calculated using the 1:1 binding model.
Inhibition of SARS-CoV-2 replication by RBD-binding mAb against spike protein
The virus inhibitory effect of mAb against Spike protein (selected by RBD-binding) was studies in a standard microneutralization assay on Vero-CCL81 cells, as described earlier (Matyushenko et al. 2021). Briefly, two-fold dilutions of antibody were prepared on DMEM/2%FBS medium in sterile U-bottom 96-well plates in a volume of 160 µL. Importantly, tested mAbs against Spike protein were diluted in a human serum sample (final concentration 10 μg/mL) to ensure the presence of human serum proteins which could potentially interfere with mAb binding to the virus. A serum sample from a COVID-19 convalescent patient was used as a positive control, while sera from a COVID-19 naive subject was used as a negative control. Each sample was assessed in duplicates. Fifty microliters of each dilution were transferred to a new 96-well plate and an equal volume of 300 TCID50 of SARS-CoV-2 was added to each well, followed by 1-h incubation at 37 °C. Then, the virus-antibody mixtures were applied to 96-well culture plates with confluent monolayer of Vero-CCL81 cells, followed by additional incubation for 1 h at 37 °C, 5% CO2. After adsorption, the inoculum was removed and the cells were covered with 100 µL of corresponding antibody dilutions. Finally, 50 µL of DMEM/2%FBS was added to each well, and the plates were incubated at 37 °C, 5% CO2. Two days later, the plates were fixed with 10% formaldehyde for 24 h at 4 °C, and the plates were subjected to cell-ELISA using polyclonal rabbit antibody specific to SARS-CoV-2 Spike RBD protein (BIOCAD, Saint Petersburg, Russia) as a primary antibody and anti-rabbit IgG HRP-conjugated antibody (Bio-Rad) as a secondary antibody. The plates were developed with 1-Step™ Ultra TMB-ELISA Substrate Solution (Thermo, USA) and the signal was read on xMark Microplate Absorbance Spectrophotometer (Bio-Rad). The MN50 titers were calculated using four-parametrical non-linear regression analysis as described in (Amanat et al. 2020).
Assessment of antiviral effect of lactoferrin in vitro
The antiviral effect of Lf was studied as described earlier (Sokolov et al. 2022a), with the exception that Vero E6 cells were used in this test. Briefly, 24-h cell monolayers in 96-well plates were infected with 100 TCID50 of SARS-CoV-2 per well, for 2 h at 37 °C, 5% CO2. After infection, the inoculum with residual virus was removed, followed by the addition of 150 µL of DMEM/2%FBS medium containing various concentrations of apo-Lf and holo-Lf. Each Lf concentration was tested in triplicate, and appropriate positive and negative controls were included. After incubation for 48 h (for B.1 Wuhan strain) or 72 h (for B.1.617.2 Delta strain), cell supernatants were harvested in 0.2 ml tubes and stored at − 70 °C until RNA extraction. Biolabmix RU-250 spin column kit (Biolabmix, Novosibirsk, Russia) was used to extract viral RNA from the collected supernatants, as well as from the viral stocks with known infectious titers. Viral load in each sample was determined by quantitative RT-PCR using Evrogen One-Tube RT-PCR SYBR kit (Moscow, Russia) with 50 × ROX as a reference dye, on Quantstudio 1 Real-Time PCR System (Thermofisher Scientific, Waltham, MA, USA). The reaction was set up as described in (Wang et al 2020b) using forward primer 5′-CAATGGTTTAACAGGCACAGG-3′ and reverse primer 5′-CTCAAGTGTCTGTGGATCACG-3′. Each RNA sample was tested in duplicate, and the average cycle threshold (Ct) value was used to calculate the virus titer from a standard curve obtained from dilutions of viral RNA with a known titer, using QuantStudio Design and Analysis Software v1.5.1.
Statistical analysis
Data presented as mean and standard deviation for measurements repeated in triplicates (unless otherwise specified). IgG antibody level presented as median and Q1-Q3, equations of linear and power regressions for antibodies levels were characterized by coefficients of determination (R2). Data of antiviral activity of Lf were analyzed by two-way ANOVA with Tukey’s post-hoc multiple analyses test.
Results
Analysis of Tf and Lf binding with human IgGs against spike protein and RBD
To test our hypothesis about molecular mimicry between RBD of Spike protein and Tf/Lf, as two homologous host ligands of TfR1, we set up several experiments of Tf or Lf binding with IgGs against Spike protein or its RBD fragment present in serum samples of study participants. First, serum levels of anti-RBD or anti-Spike IgGs were measured by indirect ELISA with corresponding antigens immobilized on solid phase (Fig. 1). Then, three variants of ELISA were performed to study the binding of these antibodies with Tf or Lf—Supplementary data (S4, Figures S2, S3, Table S1). Interaction of Lf with RBD and Spike protein, and of Spike with anti-Lf prompted to choose only one mode of ELISA with RBD or Spike protein immobilized on solid phase and IgGs detected by adding HRP-labeled anti-human Tf. We expanded the sample set of naïve, vaccinated and COVID-19-recovered individuals for studying the cross-reactivity of antibodies (“Recruiting participantsandcollecting blood samples”). Positive linear distributions of RBD- and Spike-specific IgG levels and their cross-reactivity with Tf, as measured by HRP-labeled anti-human Tf, were found with R2 values close to 1 in vaccinated and COVID-19-recovered groups (Fig. 2).
Distributions of levels of IgGs against RBD and Spike protein and Tf-binding with antibodies against RBD and Spike protein. Abscissa or ordinate designated by antigen immobilized on solid phase and a label used for detecting the level of antibodies. a—levels of IgGs against Spike protein and against RBD; b—levels of IgGs against Spike and Tf-binding with antibodies against Spike; c—levels of IgGs against RBD and Tf-binding with antibodies against RBD; d—Tf-binding with antibodies against Spike and antibodies against RBD. Vaccinated (n = 16) marked as red circles, naïve (n = 24) as green triangles, COVID-19 convalescents (n = 53) as purple rhombs
Fractionation of serum samples obtained from a naïve donor 1 day before vaccination on a sorbent with immobilized protein A showed that the IgG fraction did not contain Tf. However, Tf was detected in the IgG fractions obtained on the same column from serum samples of an individual on the 35th day after vaccination with Sputnik V and a COVID-19-recovered subject (Fig. 3a). Furthermore, IgGs obtained from sera of vaccinated and COVID-19-recovered individuals were able to bind peroxidase-labeled Lf, unlike IgGs purified from sera of naïve donors (Fig. 3b).
Co-purification of Tf with IgGs from sera of vaccinated (red) and COVID-19 convalescent (purple) persons compared with serum of naïve donor (green) sampled 1 day before vaccination. a—concentration of Tf obtained after fractionation of 3 serum samples on a column with immobilized Protein A and determined by ELISA. From 1st to 9th fraction—elution of ballast proteins, from 15th fraction start elution of IgGs; b—binding of HRP-labeled Lf with solid phase immobilized IgGs isolated from serum samples or with purified Tf as negative control
Overall, these data indicate that Spike-specific IgG antibodies can readily bind Tf and Lf, supporting our assumption of the role of the Spike, and specifically its RBD fragment, in the mimicry to host TfR ligands used in this study.
Analysis of spike protein binding with monoclonal antibodies against Lf
Since cross-reactivity between anti-Spike and anti-Lf antibodies could not be measured by the assays described above due to the direct binding of these antigens, two murine monoclonal antibodies against human Lf, namely from clones 3H5 and 3E11, were further tested. These antibodies differed by Lf-binding sites, e.g. 3E11 caused no dissociation of the complex formed by Lf with ceruloplasmin; in contrast, 3H5 antibody added to ceruloplasmin-Lf complex acted as a competitor (data not shown). Different concentrations of Lf, as well as Spike protein biotinylated either on primary amino groups (Spike-A-Bi) or on carbohydrate moiety (Spike-C-Bi) were added to 3H5 or 3E11 Lf-specific antibodies immobilized on a solid phase (Fig. 4). 3E11 antibody bound cooperatively Lf and Spike-A-Bi: the higher Lf concentration was added, the more detectable was Spike protein. In contrast, 3H5 antibody made more evident the competition between Lf and Spike protein labeled with biotin hydrazide on the carbohydrate moiety. Thus, the two anti-Lf mAbs displayed cross-reactivity both with Lf and Spike protein, however, with noticeable differences. 3H5 performed as a target for competition of the two proteins, while 3E11 could bind the complex formed by Lf and Spike protein. Quite unexpectedly, the carbohydrate-mediated labeling was more sensitive to Spike protein binding with Lf, as compared with biotin labeling via primary amines.
Competitive ELISA of Lf and Spike protein on Lf-specific antibodies (3H5 or 3E11) immobilized on solid phase. Spike protein, labeled by biotin to primary amino groups (Spike-A-biotin, 200 ng/mL—blue triangles) or to carbohydrate moiety (Spike-C-biotin, 40 ng/mL—purple squares), and different concentrations of Lf were added in wells. Subsequent staining of biotin label by HRP-streptavidin was done
Screening of TfR ligands for their ability to interfere with RBD interaction with SARS-CoV-2 neutralizing antibodies.
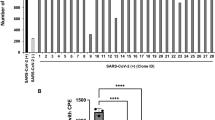
Screening among six mAbs against Spike protein for their capacity to bind RBD (Fig. 5a) and to suppress SARS-CoV-2 replication in a microneutralization assay demonstrated that S6 was the most effective in inhibiting viral replication in Vero cells, with an IC50 value close to 0.1 µg/mL (Fig. 5b). The RBD-specific mAbs S2 and S4 could partially neutralize viral activity only at the highest concentrations 0.5–1 µg/mL, suggesting different affinity of mAbs S2, S4, and S6 and/or binding to different epitopes in RBD.
Screening for RBD-binding capacity of mAbs against Spike (panel a), SARS-CoV-2 neutralization assay (panel b), competitors’ of RBD and mAbs S6 interaction (panel c), and interaction RBD with sTfR and Lf (panel d). a Spike-specific antibodies (S1-S6) screened for binding Spike and RBD immobilized on solid phase; detection by anti-mouse IgG-HRP. b SARS-CoV-2 neutralizing activity of S2, S4 and S6 mAbs. Serial dilutions of MAbs in serum of SARS-CoV-2-naïve individual were mixed with SARS-CoV-2 virus, and Vero-CCL81 cells were infected with these mixtures. The inhibitory effect of the tested antibodies was measured by cell-ELISA and expressed as % of virus inhibition compared with the control virus-infected cells using four-parametrical non-linear regression analysis. c Anti-RBD-mAb (S6), labeled with HRP, and different concentrations of sTfR (0.7–44 nM), Tf or Lf (10–640 nM), aptamer against TfR1 (50–500 nM), peptides RGD (2–120 μM) and GHAIYPRH (0.33–21 μM) were added. d TAMRA-labeled proteins run in conditions of disc-electrophoresis without detergents: 1—Fe2-Lf-TAMRA (4 mg); 2—Fe2-Lf-TAMRA (4 mg) + sTfR (4 mg); 3—sTfR (4 mg); 4—RBD-TAMRA (2 mg) + sTfR (4 mg); 5—RBD-TAMRA (2 mg); 6—Fe2Tf-TAMRA (2 mg); 7—Fe2Tf-TAMRA (2 mg) + sTfR (4 mg); 8—RBD-TAMRA (4 mg) + Fe2Lf (9 mg); 9—RBD-TAMRA (4 mg) + Fe2Lf (9 mg) + sTfR (8 mg); 10—RBD-TAMRA (4 mg) + sTfR (8 mg); 11—RBD-TAMRA (4 mg)
Interaction between S6-HRP and RBD immobilized on solid phase served as a target in the screening for interference among various ligands of TfR and sTfR itself. We assessed the binding of mAb S6-HRP to RBD in the presence of different concentrations of sTfR (0.7–44 nM), Tf or Lf (10–640 nM), aptamer against TfR1 (50–500 nM), as well as peptides RGD (2–120 μM) and GHAIYPRH (0.33–21 μM) (Fig. 5c). Importantly, sTfR showed the highest competitive activity, with IC50 value about 10 nM, while its physiological ligands i.e. Tf and Lf demonstrated virtually equal activity with IC50 about 200 nM (Fig. 5c). Involvement of structural mimicry of RBD/Spike protein to Tf and Lf was also confirmed by competitive interference of other TfR ligands in the interaction between RBD and mAb S6. Aptamer against TfR1 and peptide GHAIYPRH, as well as peptide RGD, the synthetic integrin-binding motif, are involved in the binding of Lf and Tf with TfR1. Both Tf and Lf interacted with sTfR (Fig. 5d, lines 1–2 and 6–7). Interaction of RBD with Lf was stronger (Fig. 5d, line 8–9) than interaction of RBD with sTfR (Fig. 5d, line 4–5 and 10–11).
Analysis of Lf binding to spike protein and RBD
Interaction of Lf with Spike protein has been documented already in three studies (Campione et al. 2021; Miotto et al. 2021; Cutone et al. 2022). Interaction of Lf with Spike protein prevents its interaction with anti-Lf mAbs (Fig. 4) and leads to many difficulties in assaying the cross-reactivity between antibodies raised against Spike protein or RBD and Lf. Interaction of Lf with RBD was detected by non-denaturing disc-PAGE (Fig. 6a–c). Binding of Lf with RBD was much more evident than Lf binding with Spike protein. Indeed, cationic Lf in complex with RBD migrated from stacking gel to resolving gel, while Spike protein in complex with Lf migrated from the middle of stacking gel to the space between stacking and resolving gel (Fig. 6a). Moreover, staining of Lf and RBD in Western blotting makes visible the migration of RBD from resolving to stacking gel (Fig. 6b), while using anti-Lf antibodies allowed to compare migration in electrophoresis of pure LF and its samples with RBD (Fig. 6c). Affinity of holo- and apo-Lf towards RBD was compared using RBD immobilized on CM5-chip for SPR-assay (Fig. 6d, e). Affinity towards RBD was higher for holo-Lf as judged by rate constant 4.3 × 106 M−1 × s−1 and dissociation constant 16 nM in comparison with the respective values obtained for apo-Lf i.e. 2.9 × 106 M−1 × s−1 and 23 nM (Table 2).
Interaction of Lf with RBD and Spike protein in conditions of disc-electrophoresis without detergents (panel a, b, and c) and interaction of holo-Lf and apo-Lf (4–256 nM) with RBD immobilized on CM5-chip in SPR-assay (panels d and e). a Coomassie R-250 staining of gel after running samples: 1—RBD (4 μg); 2—RBD (4 μg) + LF (18 μg); 3—RBD (4 μg) + LF (9 μg); 4—RBD (4 μg) + LF (5 μg); 5—RBD (4 μg); 6—LF (18 μg); 7—Spike (2 μg); 8—Spike (2 μg) + LF (5 μg); 9—Spike (2 μg) + LF (9 μg); 10—Spike (2 μg) + LF (18 μg). b and c Western-blotting stained with 4-chloro-1-naphtol and hydrogen peroxide after incubation with HRP-labeled anti-RBD (0.5 μg/mL S2-HRP, panel b) and with anti-Lf (0.5 μg/mL 3E11-HRP, panel c) transferred to nitrocellulose from gel after running samples: 1—RBD (2 μg) + LF (4 μg); 2—RBD (2 μg) + LF (4 μg); 3—RBD (2 μg) + LF (8 μg); 4—RBD (2 μg) + LF (16 μg); 5—RBD (2 μg); 6—empty; 7—LF (16 μg); 8—LF (8 μg); 9—LF (4 μg); 10—LF (2 μg)
Treatment of Vero E6 cells with apo-Lf and holo-Lf at concentrations 1–4 mg/mL significantly reduced SARS-CoV-2 titers, both for Wuhan and Delta lineages (Fig. 7). In all cases adding Lf at concentration 2 mg/mL substantially inhibited viral replication, and treatment with holo-Lf at this concentration resulted in the absence of detectable SARS-CoV-2 Wuhan variant (Fig. 7a). Although less pronounced reduction of Delta variant titers was seen in plates cultured in the presence of Lf, a statistically significant difference with the titer in the control wells was achieved for concentrations of 1, 2, and 4 mg/mL for both Lfs (Fig. 7b).
Lf in apo- and holo-forms inhibits SARS-CoV-2 replication in Vero E6 cells. Cells were inoculated with 100 TCID50 of B.1 Wuhan strain a or B.1.617.2 Delta strain b for 2 h, followed by the replacement of inoculum with culture medium containing either apo-Lf or holo-Lf at the indicated concentrations. Two to three days post-infection, viral load in cell supernatants was determined by quantitative RT-PCR. Data were analyzed by two-way ANOVA with Tukey’s post-hoc multiple analyses test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Discussion
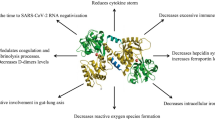
In this paper we tried to verify a hypothesis about molecular mimicry of Spike protein, especially its RBD fragment, to natural ligands of TfR, Tf and Lf, and major findings of this paper were schematically summarized in Fig. 8. On the one side, if TfR is the critical receptor for SARS-CoV-2, its natural ligand, namely Lf, seems to be a promising competitor of coronavirus cell entry; on the other side, cross-reactivity of antibodies against RBD of Spike protein with Tf and Lf can provoke severe symptoms of COVID-19 accompanied by an autoimmune response against Lf and Tf. Considering that replication cycle of SARS-CoV-2 mediated by TfR1 endocytosis can alter normal Tf-mediated influx of iron, it is not surprising that COVID-19-associated dysregulations of iron and oxygen metabolism, such as low levels of serum iron and erythropoietin accompanied by high levels of ferritin and hepcidin, can be corrected by Lf (Suriawinata and Mehta 2022). Anti-anemic and anti-hypoxic effects of Lf associated with down-regulation of IL-6 and hepcidin (Cutone et al. 2014; Rosa et al. 2017; Lepanto et al. 2019; Artym et al. 2021) and up-regulation of erythropoietin and ceruloplasmin (Pulina et al. 2010; Zakharova et al. 2012, 2018; Bonaccorsi di Patti et al. 2018; Kostevich et al. 2016; Sokolov et al. 2022b) seem to be a promising strategy for treating COVID-19 complications. It is worth noting that one of the first papers dedicated to the protective effect of Lf against SARS-CoV-2 infection also described the studies of antiviral effect of hypoxia, which decreased the content of heparan sulphate on cell surface by downregulating syndecan-1 expression via hypoxia-inducible factor-1 alpha (HIF-1α)-dependent mechanism (Prieto-Fernández et al. 2021). It cannot be excluded that antiviral effect of Lf is realized partly due to its HIF-1α-stabilizing effect (Zakharova et al. 2012).
At the beginning of this study, we did not anticipate that human antibodies against RBD and Spike protein itself would circulate in complex with Tf. However, this phenomenon explains why early attempts to detect cross-reactivity of anti-RBD or anti-Spike antibodies using either Tf on solid phase or labeled Tf were puzzling. Indeed, either reverse dependence was found between anti-RBD and anti-Spike levels and their capacity to bind Tf immobilized on solid phase (Fig. S3a, b) or, in case of labeled Tf, no dependence was observed (Fig. S3c, d).
However, when labeled anti-Tf antibody were used, we found a strong correlation between the levels of anti-RBD and anti-Spike IgG and the level of Tf bound to these antibodies (Fig. S3e–h, and Fig. 2). Importantly, in the preliminary control experiments, anti-Tf was tested for the absence of cross-reactivity with RBD and Spike protein. Indeed, the level of Tf binding in the assay of serum samples obtained from naïve donors was negligible. Considering the difficulties of detecting separately IgM and IgA, we had no aim to study the cross-reactivity with Tf of anti-RBD and anti-Spike protein belonging to each of those Ig classes, though their input in the cross-reactivity cannot be excluded. Thus, the setup of the 3rd assay accounts for possible detection of human IgA and IgM raised against RBD and Spike protein, that are cross-reactive to Tf (Fig. S2). Given that Tf binding can occur not only due to IgG, but IgA and IgM as well, the array of most points in the distribution of Tf binding above the approximating lines characterizing the anti-Spike IgG levels (Fig. 2b) and anti-RBD IgG distribution (Fig. 2c), can result from the involvement of other classes of Igs against Spike and RBD in Tf binding. TfR mediates penetration of Tf through the blood–brain barrier (Liang et al. 2018; Pardridge 2022). Since the immune response to SARS-CoV-2 induces binding of plasma Tf to anti-RBD and anti-Spike antibodies, this effect may alter iron traffic from the bloodstream into the brain. On the other hand, this may contribute to the TfR-mediated penetration of the virus through the blood–brain barrier.
Interestingly, not only human antibodies raised against RBD and Spike protein in response to SARS-CoV-2 infection or to vaccination with Sputnik V cross-reacted with human Tf and Lf (Figs. 2 and 3), but also Lf-specific mAbs 3H5 and 3E11 cross-reacted with Spike protein, as well as mAb S6 against RBD of Spike protein cross-reacted with TfR ligands (Figs. 4 and 5c). Since we observed cross-reactivity of Spike protein with mAbs 3H5 and 3E11 that were raised after immunization of mice with human Lf, we carefully checked the Spike protein for the absence of contamination with Lf. Importantly that all steps from immunizing mice to cloning hybridomas that produced anti-Lf mAbs for this study, were completed by October 2019, before the COVID-19 pandemic and the first receipt of the SARS-CoV-2 Spike protein by our laboratory.
To provide further evidence of anti-RBD and anti-Spike antibody binding with Tf and Lf, we used serum samples from naïve, vaccinated, and COVD-19-recovered donors and purified IgG on Protein A affinity sorbent. Importantly, the serum samples of naïve and vaccinated donor were obtained from the same subjects 1 day before and on the 35th day after vaccination. Tf was present only in IgG fractions purified from the sera of vaccinated and convalescent donors, and those IgGs also bound HRP-labeled Lf. IgGs obtained from the sera of naïve donors had no traces of Tf and did not bind the labeled Lf. Autoantibodies against Tf have been documented in several case-report studies of acquired anemia and myeloma-associated hypersideremia (Westerhausen and Meuret 1977; Numata et al. 1991; Alyanakian et al. 2007; Forni et al. 2008). The “gold standard” of such studies is co-purification of Tf with antibodies isolated by affinity chromatography from the patient sera (Forni et al. 2013). High concentrations of anti-Tf in autoimmune pathologies lead to dysregulation of iron metabolism, including increased levels of serum iron, Tf, ferritin, iron saturation of Tf and total serum iron-binding capacity (Alyanakian et al. 2007; Forni et al. 2008). Few of these symptoms coincide with dysregulations of iron metabolism observed in the COVID-19 patients (Suriawinata and Mehta 2022), which can be explained by much lower level of Tf-cross-reactive antibodies induced by SARS-CoV-2 infection, as compared with anti-Tf serum level in those case-report studies. However, an increase of Tf expression induced by SARS-CoV-2 infection was described (McLaughlin et al. 2020), which probably increased the risk of hypercoagulation (Tang et al. 2020b). Increased serum iron, ferritin and Tf saturation in intensive care unit patients during the 3rd–6th day of COVID-19 were documented (Bolondi et al 2020). In a pioneering study by Westerhausen and Meuret, immunosuppressive therapy in the so-called transferrin-immune complex disease only partially restored iron metabolism (Westerhausen and Meuret 1977). Extensive studies of the last decade confirmed the notion of anti-Tf reactivity of antibodies in patients with multiple sclerosis (Colomba et al. 2014).
Autoantibodies against Lf have been documented in such pathologies as anti-neutrophil cytoplasm antibodies-associated systemic vasculitis, chronic obstructive pulmonary disease, systemic lupus erythematosus, rheumatoid arthritis, ulcerative colitis, Crohn’s disease, autoimmune hepatitis (also associated with HIV-1 infections) (Peen et al. 1993; Dapsanse et al. 2001; Tan et al. 2014; Ma et al. 2020). Interestingly, antibodies cross-reacting with Lf and myeloperoxidase were reported in renal pathology associated with formation of anti-neutrophil cytoplasm autoantibodies (Esnault et al. 1994). Lf-containing immunocomplexes featured extraordinary potent proinflammatory properties against human monocytes and macrophages (Hu et al. 2017). Emergence of anti-Lf causes an increase in nitrite levels (Dapsanse et al. 2001), alteration of iron-saturation of Lf (Audrain et al. 1996), enhancement of neutrophil extracellular traps formation (NETs) (Shida et al. 2016). The latter is a well-known feature of severe COVID-19 (Ackermann et al. 2021). Effect of Lf against NETs formation is described in several studies (Okubo et al. 2016; Grigorieva et al. 2022) and this property makes it a promising remedy in mitigating the severity of COVID-19.
In ELISA with anti-Lf mAb 3E11 immobilized on solid phase, Lf and Spike protein were bound cooperatively, but the intensity of Spike binding depended on the mode of its biotinylation. When biotin was attached to primary amino groups, the interaction of Spike with mAb 3E11 was more intense than in case of biotinylated carbohydrates (Fig. 4). Meanwhile, anti-Lf mAb 3H5 immobilized on solid phase mostly bound Lf. The latter behaved as competitor of Spike protein, especially carbohydrate-biotinylated, for interaction with 3H5. It can be concluded that carbohydrate moieties of Spike protein are more important for binding with Lf than amino acids with primary amino groups. Given that the interaction of Lf with ceruloplasmin is not dissociated by 3E11, unlike 3H5, which behaves as a competitor for ceruloplasmin binding to Lf, we can assume that the interaction site of the Spike protein overlaps with ceruloplasmin-binding site of Lf. The latter was mapped in our previous studies that provided evidence of competition for binding with Lf between ceruloplasmin and DNA, lipopolysaccharide, and heparin. All these ligands bind with N-terminal polycationic site of Lf. Peptides RRRR and RKVR, corresponding to 2RRRR5 and 25RKVR28 of Lf act in the same way (Pulina et al. 2002; Sokolov et al. 2006). Involvement of N-terminal polycationic site in Lf into the interaction with ceruloplasmin was confirmed by structural studies of their complexes (Sabatucci et al. 2007; Samygina et al. 2013). Therefore, it is likely that the Spike-binding site includes the N-terminal polycationic site of Lf.
Three independent approaches were used in this study to explore the interaction of Lf with RBD of Spike protein:
-
1)
Competitive ELISA showed that Lf acts as a competitor of interaction between RBD immobilized on solid phase and HRP-labeled mAb against Spike protein (S6-HRP) which neutralizes SARS-CoV-2 infection in Vero cells (Fig. 5c);
-
2)
Lf shifted RBD band in disc-electrophoresis without detergents, and the same effect was obtained by Western blotting (Fig. 5a–c), which suggests direct binding of the two proteins;
-
3)
In SPR-assay, the interaction of RBD immobilized on CM5-chip with apo-Lf and holo-Lf was characterized by Kd ~ 23 and 16 nM, respectively (Fig. 6d, e; Table 2). Importantly, the interaction of Lf with RBD was characterized by higher affinity as compared with mAb S6 against Spike protein and anti-Lf mAb 3E11-HRP which was used for Lf detection by Western blotting (Fig. 6c). Therefore, when antibodies against Lf and RBD are produced during the immune response, the latter are of interest because they neutralized coronavirus infection, yet their interaction with antigens was weaker than their mutual interference. That is why Lf seems a prospective competitor of SARS-CoV-2 in its interaction with TfR. Such property of Lf should not be neglected, but the fact that Lf interacts with TfR has been omitted from a number of the most recent reviews concerning its functions (Mahidhara et al. 2015; Kell et al. 2020; Bartolomé et al. 2022).
Interaction of Lf with RGD-motif was well documented for TfR1 (Kawabata et al. 1999) and extracellular matrix proteins i.e., fibronectin and vitronectin (Sakamoto et al. 2006). Interestingly, 403RGD405-motif in RBD of Spike protein is missing in previous human coronavirus variants, but remains conserved in SARS-CoV-2 strains (Liu et al. 2020; Othman et al. 2022; Nassar et al. 2021). Considering the competitive effect of sTfR and synthetic RGD on the interaction of RBD with SARS-CoV-2-neutralizing mAb S6 (Fig. 5c), the importance of RGD-motif in RBD for interacting with its cellular receptor, namely TfR1, can be proposed. In view of extensive studies of TfR1 as a target for antibody-mediated cancer therapy (Candelaria et al. 2021), similar studies of anti-TfR1 potential in suppressing anti-SARS-CoV-2 activity are warranted.
Inhibition of SARS-CoV-2 infection was practically the same for apo-Lf and holo-Lf tested on B.1 Wuhan strain and B.1.617.2 Delta strain (Fig. 7). Holo-Lf in comparison with apo-Lf was more efficient only when taken in concentration 2 mg/mL in case of B.1 Wuhan strain infection (Fig. 7a). Similar efficiency of apo- and holo-Lf against infection can be interpreted at least in three ways: (1) close affinity of RBD both to apo- and holo-Lf presumes their similar efficiency; (2) both apo-Lf and holo-Lf can interact with TfR1; (3) high affinity of apo-Lf to ferric ions can result in formation of holo-Lf. Taking into account down-regulation of TfR1 by holo-Lf (Zhang et al. 2021) this form will be our choice in future studies. In contrast, apo-form LF can increase TfR1 expression via HIF-dependent pathway, which can result in undesired multiplying of the sites for viral uptake.
The details of studies demonstrating protective effects of Lf against SARS-CoV-2 in infected cells are summarized in Table 3. Taking into account that heparin is one of the ligands with the highest affinity for the polycationic site in Lf, it is not surprising that in all studies of the antiviral activity of Lf, the addition of heparin abolished its ability to inhibit SARS-CoV-2 (Prieto-Fernández et al. 2021; Mirabelli et al. 2021; Lai et al. 2022).
In their study, Mirabelli et al. performed an analysis of transcriptomic changes triggered by Lf in non-infected iAEC2 cells. Addition of 6.25 mM Lf to these cells resulted in significant up- and down-regulation of 1016 and 1023 genes, respectively. Up-regulated genes are involved in epithelial-mesenchymal transition, TNF-α signalling via NF-κB, and hypoxia (Mirabelli et al. 2021). This is an independent indication of Lf involvement in the regulation of inflammation and tolerance to hypoxia.
Summarizing the literature data and the results obtained in this study, we can conclude that antiviral effect of Lf can be mediated by several pleiotropic effects of this moonlighting protein: (1) Lf can directly interact with RBD of Spike protein with high affinity characterized by Kd from 16 nM for holo-Lf to 23 nM for apo-Lf; (2) Competition between Lf and RBD of Spike protein for binding with TfR1 can prevent TfR1-mediated endocytosis of SARS-CoV-2; (3) anti-inflammatory activity of Lf includes down-regulation of IL-6 and neutrophil extracellular trap formation; (4) Lf exhibits anti-hypoxic and anti-anaemic effects. All these effects of Lf seem like promising approaches to reducing the severity of COVID-19 and post-COVID syndrome.
References
Ackermann M, Anders H-J, Bilyy R et al (2021) Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ 28:3125–3139. https://doi.org/10.1038/s41418-021-00805-z
Aleem A, Akbar Samad AB, Slenker AK (2022) Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In: StatPearls. Treasure Island (FL): StatPearls Publishing; October 10, 2022.
Alkhateeb AA, Buckett PD, Gardeck AM et al (2015) The small molecule ferristatin II induces hepatic hepcidin expression in vivo and in vitro. Am J Physiol Gastrointest Liver Physiol 308:G1019–G1026. https://doi.org/10.1152/ajpgi.00324.2014
Alyanakian MA, Taes Y, Bensaïd M et al (2007) Monoclonal immunoglobulin with antitransferrin activity: a rare cause of hypersideremia with increased transferrin saturation. Blood 109:359–361. https://doi.org/10.1182/blood-2006-05-023762
Amanat F, White KM, Miorin L et al (2020) An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol 58:e108. https://doi.org/10.1002/cpmc.108
Anderson NL, Nance SL, Pearson TW et al (1982) Specific antiserum staining of two-dimensional electrophoretic patterns of human plasma proteins immobilized on nitricellulose. Electrophoresis 3:135–142
Artym J, Zimecki M, Kruzel ML (2021) Lactoferrin for prevention and treatment of anemia and inflammation in pregnant women: a comprehensive review. Biomedicines 9:898. https://doi.org/10.3390/biomedicines9080898
Audrain MA, Gourbil A, Muller JY, Esnault LM (1996) Anti-lactoferrin autoantibodies: relation between epitopes and iron-binding domain. J Autoimmun 9:569–574. https://doi.org/10.1006/jaut.1996.0076
Ayo A, Laakkonen P (2021) Peptide-based strategies for targeted tumor treatment and imaging. Pharmaceutics 13:481. https://doi.org/10.3390/pharmaceutics13040481
Baker HM, Baker EN (2004) Lactoferrin and iron: structural and dynamic aspects of binding and release. Biometals 17:209–216. https://doi.org/10.1023/b:biom.0000027694.40260.70
Bartolomé F, Rosa L, Valenti P et al (2022) Lactoferrin as immune-enhancement strategy for SARS-CoV-2 infection in Alzheimer’s disease patients. Front Immunol 13:878201. https://doi.org/10.3389/fimmu.2022.878201
Bolondi G, Russo E, Gamberini E et al (2020) Iron metabolism and lymphocyte characterisation during Covid-19 infection in ICU patients: an observational cohort study. World J Emerg Surg 15:41. https://doi.org/10.1186/s13017-020-00323-2
Bonaccorsi di Patti MC, Cutone A, Polticelli F et al (2018) The ferroportin-ceruloplasmin system and the mammalian iron homeostasis machine: regulatory pathways and the role of lactoferrin. Biometals 31:399–414. https://doi.org/10.1007/s10534-018-0087-5
Britigan BE, Serody JS, Cohen MS (1994) The role of lactoferrin as an anti-inflammatory molecule. Adv Exp Med Biol 357:143–156. https://doi.org/10.1007/978-1-4615-2548-6_14
Brock JH (1980) Lactoferrin in human milk: its role in iron absorption and protection against enteric infection in the newborn infant. Arch Dis Child 55:417–421. https://doi.org/10.1136/adc.55.6.417
Brown JX, Buckett PD, Wessling-Resnick M (2004) Identification of small molecule inhibitors that distinguish between non-transferrin bound iron uptake and transferrin-mediated iron transport. Chem Biol 11:407–416. https://doi.org/10.1016/j.chembiol.2004.02.016
Byrne SL, Buckett PD, Kim J et al (2013) Ferristatin II promotes degradation of transferrin receptor-1 in vitro and in vivo. PLoS ONE 8:e70199. https://doi.org/10.1371/journal.pone.0070199
Campione E, Lanna C, Cosio T et al (2021) Lactoferrin against SARS-CoV-2: in vitro and in silico evidences. Front Pharmacol 12:666600. https://doi.org/10.3389/fphar.2021.666600
Candelaria PV, Leoh LS, Penichet ML, Daniels-Wells TR (2021) Antibodies targeting the transferrin receptor 1 (TfR1) as direct anti-cancer agents. Front Immunol 12:607692. https://doi.org/10.3389/fimmu.2021.607692
Cantuti-Castelvetri L, Ojha R, Pedro LD et al (2020) Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370:856–860. https://doi.org/10.1126/science.abd2985
Cavezzi A, Menicagli R, Troiani E, Corrao S (2022) COVID-19, cation dysmetabolism, sialic acid, CD147, ACE2, viroporins, hepcidin and ferroptosis: a possible unifying hypothesis. Research 11:102. https://doi.org/10.12688/f1000research.108667.2
Colomba P, Fontana S, Salemi G et al (2014) Identification of biomarkers in cerebrospinal fluid and serum of multiple sclerosis patients by immunoproteomics approach. Int J Mol Sci 15:23269–23282. https://doi.org/10.3390/ijms151223269
Cutone A, Frioni A, Berlutti F et al (2014) Lactoferrin prevents LPS-induced decrease of the iron exporter ferroportin in human monocytes/macrophages. Biometals 27:807–813. https://doi.org/10.1007/s10534-014-9742-7
Cutone A, Rosa L, Bonaccorsi di Patti MC et al (2022) Lactoferrin binding to Sars-CoV-2 Spike glycoprotein protects host from infection, inflammation and iron dysregulation. Res Sq. https://doi.org/10.21203/rs.3.rs-1605740/v1
Dakal TC (2021) SARS-CoV-2 attachment to host cells is possibly mediated via RGD-integrin interaction in a calcium-dependent manner and suggests pulmonary EDTA chelation therapy as a novel treatment for COVID 19. Immunobiology 226:152021. https://doi.org/10.1016/j.imbio.2020.152021
Daly JL, Simonetti B, Klein K et al (2020) Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370:861–865. https://doi.org/10.1126/science.abd3072
Dapsanse V, Defer MC, Follézou JY et al (2001) Differential pattern in circulating nitrogen derivatives, lactoferrin, and anti-lactoferrin antibodies in HIV type 1 and HIV type 2 infections. AIDS Res Hum Retrovir 17:1041–1045. https://doi.org/10.1089/088922201300343726
Davis BJ (1964) Disc electrophoresis. II. Method and application to human serum proteins. Ann N Y Acad Sci 121:404–427
Dewan A, Jani JP, Patel JS et al (1988) Benzidine and its acetylated metabolites in the urine of workers exposed to direct black 38. Arch Environ Health 43:269–272. https://doi.org/10.1080/00039896.1988.10545948
Dubljevic V, Sali A, Goding JW (1999) A conserved RGD (Arg-Gly-Asp) motif in the transferrin receptor is required for binding to transferrin. Biochem J 341:11–14
Elzoghby AO, Abdelmoneem MA, Hassanin IA et al (2020) Lactoferrin, a multi-functional glycoprotein: Active therapeutic, drug nanocarrier & targeting ligand. Biomaterials 263:120355. https://doi.org/10.1016/j.biomaterials.2020.120355
Esnault VL, Short AK, Audrain MA et al (1994) Autoantibodies to lactoferrin and histone in systemic vasculitis identified by anti-myeloperoxidase solid phase assays. Kidney Int 46(1):153–160. https://doi.org/10.1038/ki.1994.254
Fling SP, Gregerson DS (1986) Peptide and protein molecular weigth determination by electrophoresis using a high-molarity tris buffer system without urea. Anal Biochem 155:83–88
Forni GL, Girelli D, Lamagna M et al (2008) Acquired iron overload associated with antitransferrin monoclonal immunoglobulin: a case report. Am J Hematol 83:932–934. https://doi.org/10.1002/ajh.21297
Forni GL, Pinto V, Musso M et al (2013) Transferrin-immune complex disease: a potentially overlooked gammopathy mediated by IgM and IgG. Am J Hematol 88:1045–1049. https://doi.org/10.1002/ajh.23558
Godínez-Chaparro B, Guzmán-Mejía F, Drago-Serrano ME (2021) Lactoferrin and its potential impact for the relief of pain: a preclinical approach. Pharmaceuticals 14:868. https://doi.org/10.3390/ph14090868
Grigorieva DV, Gorudko IV, Grudinina NA et al (2022) Lactoferrin modified by hypohalous acids: partial loss in activation of human neutrophils. Int J Biol Macromol 195:30–40. https://doi.org/10.1016/j.ijbiomac.2021.11.165
Hu L, Hu X, Long K et al (2017) Extraordinarily potent proinflammatory properties of lactoferrin-containing immunocomplexes against human monocytes and macrophages. Sci Rep 7:4230. https://doi.org/10.1038/s41598-017-04275-7
Hu Y, Meng X, Zhang F et al (2021) The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerg Microbes Infect 10:317–330. https://doi.org/10.1080/22221751.2021.1888660
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. https://doi.org/10.1038/s41580-021-00418-x
Jennifer B, Berg V, Modak M et al (2020) Transferrin receptor 1 is a cellular receptor for human heme-albumin. Commun Biol 3:621. https://doi.org/10.1038/s42003-020-01294-5
Kanevsky VYu, Pozdnyakova LP, Katukov VYu, Severin SE (1997) Isolation of the transferrin receptor from human placenta. Biochem Mol Biol Int 42:309–314. https://doi.org/10.1080/15216549700202701
Kawabata H, Yang R, Hirama T et al (1999) Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J Biol Chem 274:20826–20832. https://doi.org/10.1074/jbc.274.30.20826
Kawakami H, Dosako S, Lönnerdal B (1990) Iron uptake from transferrin and lactoferrin by rat intestinal brush-border membrane vesicles. Am J Physiol 258:G535–G541. https://doi.org/10.1152/ajpgi.1990.258.4.G535
Kell DB, Heyden EL, Pretorius E (2020) The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front Immunol 11:1221. https://doi.org/10.3389/fimmu.2020.01221
Kostevich VA, Sokolov AV, Kozlov SO et al (2016) Functional link between ferroxidase activity of ceruloplasmin and protective effect of apo-lactoferrin: studying rats kept on a silver chloride diet. Biometals 29:691–704. https://doi.org/10.1007/s10534-016-9944-2
Kronstein-Wiedemann R, Stadtmüller M, Traikov S et al (2022) SARS-CoV-2 infects red blood cell progenitors and dysregulates hemoglobin and iron metabolism. Stem Cell Rev Rep 18:1809–1821. https://doi.org/10.1007/s12015-021-10322-8
Kumar S, Sheokand N, Mhadeshwar MA et al (2012) Characterization of glyceraldehyde-3-phosphate dehydrogenase as a novel transferrin receptor. Int J Biochem Cell Biol 44:189–199. https://doi.org/10.1016/j.biocel.2011.10.016
Lai X, Yu Y, Xian W et al (2022) Identified human breast milk compositions effectively inhibit SARS-CoV-2 and variants infection and replication. iScience 25:104136. https://doi.org/10.1016/j.isci.2022.104136
Lebrón JA, West AP Jr, Bjorkman PJ (1999) The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol 294:239–245. https://doi.org/10.1006/jmbi.1999.3252
Lepanto MS, Rosa L, Paesano R et al (2019) Lactoferrin in aseptic and septic inflammation. Molecules 24:1323. https://doi.org/10.3390/molecules24071323
Li YQ, Guo C (2021) A review on lactoferrin and central nervous system diseases. Cells 10:1810. https://doi.org/10.3390/cells10071810
Liang M, Gao C, Wang Y et al (2018) Enhanced blood-brain barrier penetration and glioma therapy mediated by T7 peptide-modified low-density lipoprotein particles. Drug Deliv 25:1652–1663. https://doi.org/10.1080/10717544.2018.1494223
Liu Z, Xiao X, Wei X et al (2020) Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. https://doi.org/10.1002/jmv.25726
Luck AN, Mason AB (2013) Structure and dynamics of drug carriers and their interaction with cellular receptors: focus on serum transferrin. Adv Drug Deliv Rev. 65(8):1012–1019. https://doi.org/10.1016/j.addr.2012.11.001
Ma A, Wen L, Yin J et al (2020) Serum levels of autoantibodies against extracellular antigens and neutrophil granule proteins increase in patients with COPD compared to non-COPD smokers. Int J Chron Obstruct Pulmon Dis 15:189–200. https://doi.org/10.2147/COPD.S235903
Mahidhara G, Kanwar RK, Roy K, Kanwar JR (2015) Oral administration of iron-saturated bovine lactoferrin-loaded ceramic nanocapsules for breast cancer therapy and influence on iron and calcium metabolism. Int J Nanomed 10:4081–4098. https://doi.org/10.2147/IJN.S75877
Makowski L, Olson-Sidford W (2021) Biological and clinical consequences of integrin binding via a rogue RGD motif in the SARS CoV-2 spike protein. Viruses 13:146. https://doi.org/10.3390/v13020146
Malhotra H, Kumar M, Chauhan AS et al (2019) Moonlighting protein glyceraldehyde-3-phosphate dehydrogenase: a cellular rapid-response molecule for maintenance of iron homeostasis in hypoxia. Cell Physiol Biochem 52:517–531. https://doi.org/10.33594/000000037
Masson PL, Heremans JF, Dive CH (1966) Studies on lactoferrin, an iron-binding protein common to many external secretions. Clin Chim Acta 14:735–739. https://doi.org/10.1016/0009-8981(66)90004-0
Masson PL (1970) In: Arscia SA (ed.) La lactoferrine. proteine des secretions externes et des leucocytes neutrophiles, Brussels
Matyushenko V, Isakova-Sivak I, Kudryavtsev I et al (2021) Detection of IFNγ-secreting CD4+ and CD8+ memory T cells in COVID-19 convalescents after stimulation of peripheral blood mononuclear cells with live SARS-CoV-2. Viruses 13:1490. https://doi.org/10.3390/v13081490
McLaughlin KM, Bechtel M, Bojkova D et al (2020) COVID-19-related coagulopathy-is transferrin a missing link? Diagnostics 10:539. https://doi.org/10.3390/diagnostics10080539
Miotto M, Di Rienzo L, Bò L et al (2021) Molecular mechanisms behind anti SARS-CoV-2 action of lactoferrin. Front Mol Biosci 8:607443. https://doi.org/10.3389/fmolb.2021.607443
Mirabelli C, Wotring JW, Zhang CJ et al (2021) Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19. Proc Natl Acad Sci USA 118:e2105815118. https://doi.org/10.1073/pnas.2105815118
Naidu SAG, Clemens RA, Naidu AS (2022) SARS-CoV-2 infection dysregulates host iron (Fe)-redox homeostasis (Fe-R-H): role of Fe-redox regulators, ferroptosis inhibitors, anticoagulants, and iron-chelators in COVID-19 control. J Diet Suppl. https://doi.org/10.1080/19390211.2022.2075072
Nassar A, Ibrahim IM, Amin FG et al (2021) A review of human coronaviruses’ receptors: the host-cell targets for the crown bearing viruses. Molecules 26:6455. https://doi.org/10.3390/molecules26216455
Norris EG, Pan XS, Hocking DC (2022) Receptor binding domain of SARS-CoV-2 is a functional αv-integrin agonist. bioRxiv. https://doi.org/10.1101/2022.04.11.487882
Numata Y, Tanioka F, Yoshida K et al (1991) Ascertainment of IgA1 (kappa)-transferrin complex in a case of multiple myeloma associated with hypersiderinemia. Jpn J Med 30:498–503. https://doi.org/10.2169/internalmedicine1962.30.498
Okubo K, Kamiya M, Urano Y et al (2016) Lactoferrin suppresses neutrophil extracellular traps release in inflammation. EBioMedicine 10:204–215. https://doi.org/10.1016/j.ebiom.2016.07.012
Othman H, Messaoud HB, Khamessi O et al (2022) SARS-CoV-2 spike protein unlikely to bind to integrins via the Arg-Gly-Asp (RGD) motif of the receptor binding domain: evidence from structural analysis and microscale accelerated molecular dynamics. Front Mol Biosci 9:834857. https://doi.org/10.3389/fmolb.2022.834857
Pandey S (2010) Hybridoma technology for production of monoclonal antibodies. Int J Pharm Sci Res 1:88–94
Pardridge WM (2022) Kinetics of blood-brain barrier transport of monoclonal antibodies targeting the insulin receptor and the transferrin receptor. Pharmaceuticals 15:3. https://doi.org/10.3390/ph15010003
Peen E, Almer S, Bodemar G et al (1993) Anti-lactoferrin antibodies and other types of ANCA in ulcerative colitis, primary sclerosing cholangitis, and Crohn’s disease. Gut 34:56–62. https://doi.org/10.1136/gut.34.1.56
Prieto-Fernández E, Egia-Mendikute L, Vila-Vecilla L et al (2021) Hypoxia reduces cell attachment of SARS-CoV-2 spike protein by modulating the expression of ACE2, neuropilin-1, syndecan-1 and cellular heparan sulfate. Emerg Microbes Infect 10:1065–1076. https://doi.org/10.1080/22221751.2021.1932607
Prokofyev A, Gershovich P, Strelkova A, et al (2021) AAV5-based vaccine for induction of specific immunity to SARS-CoV-2 virus and/or prevention of SARS-CoV-2-induced coronavirus infection. Patent RU 2761879 C1 Published 13.12.2021
Pulina MO, Zakharova ET, Sokolov AV et al (2002) Studies of the ceruloplasmin-lactoferrin complex. Biochem Cell Biol 80:35–39. https://doi.org/10.1139/o01-206
Pulina MO, Sokolov AV, Zakharova ET et al (2010) Effect of lactoferrin on consequences of acute experimental hemorrhagic anemia in rats. Bull Exp Biol Med 149:219–222. https://doi.org/10.1007/s10517-010-0911-6
Rawat P, Kumar S, Sheokand N et al (2012) The multifunctional glycolytic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a novel macrophage lactoferrin receptor. Biochem Cell Biol 90:329–338. https://doi.org/10.1139/o11-058
Römer M, Eichner J, Metzger U et al (2014) Cross-platform toxicogenomics for the prediction of non-genotoxic hepatocarcinogenesis in rat. PLoS ONE 9:e97640. https://doi.org/10.1371/journal.pone.0097640
Rosa L, Cutone A, Lepanto MS et al (2017) Lactoferrin: a natural glycoprotein involved in iron and inflammatory homeostasis. Int J Mol Sci 18:1985. https://doi.org/10.3390/ijms18091985
Sabatucci A, Angelucci CB, Maccarrone M et al (2007) Structural characterization of the ceruloplasmin: lactoferrin complex in solution. J Mol Biol 371:1038–1046. https://doi.org/10.1016/j.jmb.2007.05.089
Sakamoto K, Ito Y, Mori T, Sugimura K (2006) Interaction of human lactoferrin with cell adhesion molecules through RGD motif elucidated by lactoferrin-binding epitopes. J Biol Chem 281:24472–24478. https://doi.org/10.1074/jbc.M604974200
Sakamoto S, Kawabata H, Masuda T et al (2015) H-ferritin is preferentially incorporated by human erythroid cells through transferrin receptor 1 in a threshold-dependent manner. PLoS ONE 10:e0139915. https://doi.org/10.1371/journal.pone.0139915
Salaris C, Scarpa M, Elli M et al (2021) Protective effects of lactoferrin against SARS-CoV-2 infection in vitro. Nutrients 13:328. https://doi.org/10.3390/nu13020328
Samygina VR, Sokolov AV, Bourenkov G et al (2013) Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins. PLoS ONE 8:e67145. https://doi.org/10.1371/journal.pone.0067145
Scialo F, Daniele A, Amato F et al (2020) ACE2: the major cell entry receptor for SARS-CoV-2. Lung 198:867–877. https://doi.org/10.1007/s00408-020-00408-4
Seligman PA, Allen RH (1987) Isolation of transferrin receptor from human placenta. Methods Enzymol 147:239–247. https://doi.org/10.1016/0076-6879(87)47114-0
Semak I, Budzevich A, Maliushkova E et al (2019) Development of dairy herd of transgenic goats as biofactory for large-scale production of biologically active recombinant human lactoferrin. Transgenic Res 28:465–478. https://doi.org/10.1007/s11248-019-00165-y
Shang J, Ye G, Shi K et al (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. https://doi.org/10.1038/s41586-020-2179-y
Shida H, Nakazawa D, Tateyama Y et al (2016) The presence of anti-lactoferrin antibodies in a subgroup of eosinophilic granulomatosis with polyangiitis patients and their possible contribution to enhancement of neutrophil extracellular trap formation. Front Immunol 7:636. https://doi.org/10.3389/fimmu.2016.00636
Sokolov AV, Pulina MO, Zakharova ET et al (2006) Identification and isolation from breast milk of ceruloplasmin-lactoferrin complex. Biochemistry 71:160–166. https://doi.org/10.1134/s0006297906020076
Sokolov AV, Voynova IV, Kostevich VA et al (2017) Comparison of Interaction between Ceruloplasmin and Lactoferrin/Transferrin: to Bind or Not to Bind. Biochemistry (Mosc). 82:1073–1078. https://doi.org/10.1134/S0006297917090115
Sokolov A, Isakova-Sivak I, Grudinina N et al (2022a) Ferristatin II efficiently inhibits SARS-CoV-2 replication in vero cells. Viruses 14:317. https://doi.org/10.3390/v14020317
Sokolov AV, Dubrovskaya NM, Kostevich VA et al (2022b) Lactoferrin induces erythropoietin synthesis and rescues cognitive functions in the offspring of rats subjected to prenatal hypoxia. Nutrients 14:1399. https://doi.org/10.3390/nu14071399
Suriawinata E, Mehta KJ (2022) Iron and iron-related proteins in COVID-19. Clin Exp Med 2022:1–23. https://doi.org/10.1007/s10238-022-00851-y
Suzuki YA, Lopez V, Lönnerdal B (2005) Mammalian lactoferrin receptors: structure and function. Cell Mol Life Sci 62:2560–2575. https://doi.org/10.1007/s00018-005-5371-1
Tan L, Zhang Y, Peng W et al (2014) Detection of anti-lactoferrin antibodies and anti-myeloperoxidase antibodies in autoimmune hepatitis: a retrospective study. J Immunoassay Immunochem 35:388–397. https://doi.org/10.1080/15321819.2013.879450
Tang X, Yang M, Duan Z et al (2020a) Transferrin receptor is another receptor for SARS-CoV-2 entry. BioRxiv. https://doi.org/10.1101/2020a.10.23.350348
Tang X, Zhang Z, Fang M et al (2020b) Transferrin plays a central role in coagulation balance by interacting with clotting factors. Cell Res 30:119–132. https://doi.org/10.1038/s41422-019-0260-6
Turkewitz AP, Amatruda JF, Borhani D et al (1988) A high yield purification of the human transferrin receptor and properties of its major extracellular fragment. J Biol Chem 263:8318–8325
Wang K, Chen W, Zhang Z et al (2020a) CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Sig Transduct Target Ther 5:283. https://doi.org/10.1038/s41392-020-00426-x
Wang M, Cao R, Zhang L et al (2020b) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. https://doi.org/10.1038/s41422-020-0282-0
Wang S, Qiu Z, Hou Y et al (2021) AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31:126–140. https://doi.org/10.1038/s41422-020-00460-y
Ward JL, Torres-Gonzalez M, Ammons MCB (2022) The influence of viral infections on iron homeostasis and the potential for lactoferrin as a therapeutic in the age of the SARS-CoV-2 pandemic. Nutrients 14:3090. https://doi.org/10.3390/nu14153090
Wei C, Wan L, Yan Q et al (2020) HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab 2:1391–1400. https://doi.org/10.1038/s42255-020-00324-0
Westerhausen M, Meuret G (1977) Transferrin-immune complex disease. Acta Haematol 57:96–101. https://doi.org/10.1159/000207865
Wotring JW, Fursmidt R, Ward L, Sexton JZ (2022) Evaluating the in vitro efficacy of bovine lactoferrin products against SARS-CoV-2 variants of concern. J Dairy Sci 105:2791–2802. https://doi.org/10.3168/jds.2021-21247
Xu G, Liu R, Zak O, Aisen P, Chance MR (2005) Structural allostery and binding of the transferrin*receptor complex. Mol Cell Proteomics 4:1959–1967. https://doi.org/10.1074/mcp.M500095-MCP200
Zakharova ET, Kostevich VA, Sokolov AV, Vasilyev VB (2012) Human apo-lactoferrin as a physiological mimetic of hypoxia stabilizes hypoxia-inducible factor-1 alpha. Biometals 25:1247–1259. https://doi.org/10.1007/s10534-012-9586-y
Zakharova ET, Sokolov AV, Pavlichenko NN et al (2018) Erythropoietin and Nrf2: key factors in the neuroprotection provided by apo-lactoferrin. Biometals 31:425–443. https://doi.org/10.1007/s10534-018-0111-9
Zhang Q, Chen CZ, Swaroop M et al (2020) Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov 6:80. https://doi.org/10.1038/s41421-020-00222-5
Zhang Z, Lu M, Chen C et al (2021) Holo-lactoferrin: the link between ferroptosis and radiotherapy in triple-negative breast cancer. Theranostics 11:3167–3182. https://doi.org/10.7150/thno.52028
Zhang H, Jin C, Zhang L et al (2022) CD71-specific aptamer conjugated with monomethyl auristatin E for the treatment of uveal melanoma. ACS Appl Mater Interfaces 14:32–40. https://doi.org/10.1021/acsami.1c13980
Zhou C, Chen Y, Ji Y et al (2020) Increased serum levels of hepcidin and ferritin are associated with severity of COVID-19. Med Sci Monit 26:e926178. https://doi.org/10.12659/MSM.926178
Acknowledgements
We thank Dr. Alexandra Rak for her help in maintaining continuous cell lines. We also thank all participants who donated blood and placenta tissue for this study.
Funding
This work was supported by State assignment of the Federal State Budgetary Institution “Research Institute of Experimental Medicine” 0557-2019-0009 and FGWG-2022-0001. Equipment of the Shared Core Facilities “Human microbiome” at the Institute of Experimental Medicine was exploited upon synthesis of peptides for this research.
Author information
Authors and Affiliations
Contributions
Conceptualization: AVS, INIS; Methodology: AVS, INIS, DAM, VAK, NPG, NAG, NNK; Formal analysis and investigation: AVS, INIS, DAM, VAK, NPG, AYE, VAM, YMB; Validation: YMB, YAZ; Writing—original draft preparation: AVS, INIS; Writing—review and editing: INIS, VBV; Funding acquisition: INIS; Resources: ASK, IVS, AIB; Supervision: INIS, LGR, VBV.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sokolov, A.V., Isakova-Sivak, I.N., Mezhenskaya, D.A. et al. Molecular mimicry of the receptor-binding domain of the SARS-CoV-2 spike protein: from the interaction of spike-specific antibodies with transferrin and lactoferrin to the antiviral effects of human recombinant lactoferrin. Biometals 36, 437–462 (2023). https://doi.org/10.1007/s10534-022-00458-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-022-00458-6