Abstract
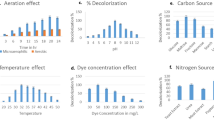
Soil samples collected from dye contaminated sites of Vatva, Gujarat, India were studied for the screening and isolation of organisms capable of decolourizing textile dyes. The most efficient isolate, which showed decolourization zone of 48 mm on 300 ppm Reactive Red BS (C.I.111) containing plate, was identified as Pseudomonas aeruginosa. Reactive Red BS (C.I.111) was used as a model dye for the study. The isolated culture exhibited 91% decolourization of 300 ppm dye within 5.5 h over a wide pH range from 5.0 to 10.5 and temperature ranging from 30 to 40°C. The culture was able to decolourize more than 91% of Reactive Red BS under static conditions in presence of either glucose, peptone or yeast extract. Addition of 300 ppm of Reactive Red BS, in each step, in ongoing dye decolourization flask, gave more than 90% decolourization within 2 h corresponding to 136 mg l−1 h−1 dye removal rate. The isolate had the ability to decolourize six different reactive dyes tested as well as the actual dye manufacturing industry’s effluent. The degradation of the dye was confirmed by HPTLC.






Similar content being viewed by others
References
Bhatt N, Patel KC, Keharia H, Madamwar D (2005) Decolorization of diazo-dye Reactive Blue 172 by Pseudomonas aeruginosa NBAR12. J Basic Microbiol 45(6):407–418
Biolog (2001) Instructions for use of the Biolog GP2 and GN2 Microplates™. Biolog INC, Hayward
Chakraborty S, Purkait MK, DasGupta S, De S, Basu JK (2003) Nanofiltration of textile plant effluent for colour removal and reduction in COD. Sep Purif Technol 31(2):141–151
Chang JS, Lin YC (2000) Fed-batch bioreactor strategies for microbial decolourization of azo dye using a Pseudomonas luteola strain. Biotechnol Progress 16:979–985
Chen BY (2002) Understanding decolorization characteristics of reactive azo dyes by Pseudomonas luteola: toxicity and kinetics. Process Biochem 38(3):437–446
Chen KC, Huang WT, Wu JY, Houng JY (1999) Microbial decolorization of azo dyes by Proteus mirabilis. J Ind Microbiol Biotechnol 23(1):686–690
Chen KC, Wu JY, Liou DJ, Hwang SCJ (2003) Decolorization of the textile dyes by newly isolated bacterial strains. J Biotechnol 101(1):57–68
Clarke GM (1994) Statistics and experimental design—an introduction for biologists and biochemists, 3rd edn. Edward Arnold, London, pp 164–174
Dave SR, Dave RH (2009) Isolation and characterization of Bacillus thuringiensis for Acid red 119 dye decolourization. Biresour Technol 100:249–253
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Georgiou D, Aivazidis A, Hatiras J, Gimouhopoulos K (2003) Treatment of cotton textile wastewater using lime and ferrous sulfate. Water Res 37(9):2248–2250
Gottlieb A, Shaw C, Smith A, Wheatley A, Forsythe S (2003) The toxicity of textile reactive azo dyes after hydrolysis and decolourization. J Biotechnol 101(1):49–56
Guo J, Zhou J, Wang D, Yang J, Li Z (2008) The new incorporation bio-treatment technology of bromoamine acid and azo dyes wastewaters under high-salt conditions. Biodegradation 19(1):93–98
Hu TL (1994) Decolourization of reactive azo dyes by transformation with Pseudomonas luteola. Bioresour Technol 49(1):47–51
Hu TL (2001) Kinetics of azoreductase and assessment of toxicity of metabolic products from azo dyes by Pseudomonas luteola. Water Sci Technol 43(2):261–269
Khehra M, Saini H, Sharma D, Chadha B, Chimni S (2005) Decolorization of various azo dyes by bacterial consortium. Dyes Pigments 67(1):55–61
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirk RE, Othmer DF (1993) Dyes, environmental chemistry. In: Howe-Grant M (ed) Kirk-Othmer encyclopaedia of chemical technology, vol 8, 4th edn. Wiley Interscience Publication, New York, pp 753–773
Kumar V, Wati L, FitzGibbon F, Nigam P, Banat IM, Singh D, Marchant R (1997) Bioremediation and decolorization of anaerobically digested distillery spent wash. Biotechnol Lett 19(4):311–314
Mali PL, Mahajan MM, Patil DP, Kulkarni MV (2000) Biodecolourisation of members of triphenylmethane and azo group of dyes. J Sci Ind Res 59:221–224
McMullan G, Meehan C, Conneely A, Kirby N, Robinson T, Nigam P, Banat IM, Marchant R, Smyth WF (2001) Microbial decolourisation and degradation of textile dyes. Appl Microbiol Biotechnol 56:81–87
Mielgo I, Moreira MT, Feijoo G, Lema JM (2002) Biodegradation of a polymeric dye in a pulsed bed bioreactor by immobilised Phanerochaete chrysosporium. Water Res 36(7):1896–1901
Moosvi S, Keharia H, Madamwar D (2005) Decolourization of textile dye reactive violet 5 by a newly isolated bacterial consortium RVM 11.1. World J Microbiol Biotechnol 21:667–672
Papić S, Koprivanac N, Lončarić Božić A (2000) Removal of reactive dyes from wastewater using Fe(III) coagulant. Color Technol 116(11):352–358
Pavan FA, Mazzocato AC, Gushikem Y (2008) Removal of methylene blue dye from aqueous solutions by adsorption using yellow passion fruit peel as adsorbent. Bioresour Technol 99(8):3162–3165
Ramsay JA, Nguyen T (2002) Decoloration of textile dyes by Trametes versicolor and its effect on dye toxicity. Biotechnol Lett 24(21):1757–1761
Robinson T, McMullan G, Marchant R, Nigam P (2001) Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour Technol 77(3):247–255
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sandhya S, Padmavathy S, Swaminathan K, Subrahmanyam YV, Kaul SN (2005) Microaerophilic-aerobic sequential batch reactor for treatment of azo dyes containing simulated wastewater. Process Biochem 40(2):885–890
Sandhya S, Sarayu K, Uma B, Swaminathan K (2008) Decolorizing kinetics of a recombinant Escherichia coli SS125 strain harbouring azoreductase gene from Bacillus latrosporus RRK1. Bioresour Technol 99(7):2187–2191
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56(1–2):69–80
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Acknowledgments
We are thankful to Ganesh Dye Chem. dye manufacturing industry, Ahmedabad for providing the dye samples, industrial effluent and waste.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sheth, N.T., Dave, S.R. Optimisation for enhanced decolourization and degradation of Reactive Red BS C.I. 111 by Pseudomonas aeruginosa NGKCTS. Biodegradation 20, 827–836 (2009). https://doi.org/10.1007/s10532-009-9270-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-009-9270-2