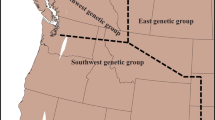
Abstract
Hybridization creates unique allele combinations which can facilitate the evolution of invasiveness. Frequent interspecific hybridization between the Siberian elm, Ulmus pumila, and native elm species has been detected in the Midwestern United States, Italy and Spain. However, Ulmus pumila also occurs in the western United States and Argentina, regions where no native elm species capable of hybridizing with it occurs. We examined whether inter- or intraspecific hybridization could be detected in these regions. Nuclear markers and the program STRUCTURE helped detect interspecific hybridization and determine the population genetic structure in both the native and the two non-native ranges. Chloroplast markers identified sources of introduction into these two non-native ranges. No significant interspecific hybridization was detected between U. pumila and U. rubra in the western United States or between U. pumila and U. minor in Argentina and vice versa. However, the genetic findings supported the presence of intraspecific hybridization and high levels of genetic diversity in both non-native ranges. The evidence presented for intraspecific hybridization in the current study, combined with reports of interspecific hybridization from previous studies, identifies elm as a genus where both inter- and intraspecific hybridization may occur and help maintain high levels of genetic diversity potentially associated with invasiveness.




Similar content being viewed by others
References
Abbott RJ, Brennan AC (2014) Altitudinal gradients, plant hybrid zones and evolutionary novelty. Philos Trans R Soc Lond B Biol Sci 369:20130346
Bertolasi B, Leonarduzzi C, Piotti A, Leonardi S, Zago L, Gui L, Gorian F, Vanetti I, Binelli G (2015) A last stand in the Po valley: genetic structure and gene flow patterns in Ulmus minor and U. pumila. Ann Bot 115:683–692
Blair AC, Hufbauer RA (2010) Hybridization and invasion: one of North America’s most devastating invasive plants shows evidence for a history of interspecific hybridization. Evol Appl 3:40–51
Bleeker W, Schmitz U, Ristow M (2007) Interspecific hybridisation between alien and native plant species in Germany and its consequences for native biodiversity. Biol Conserv 137:248–253
Brunet J, Zalapa JE, Peccori F, Santini A (2013) Hybridization and introgression between exotic Siberian elm, Ulmus pumila, and the native Field elm, U. minor, in Italy. Biol Invasions 15:2717–2730
Cabra-Rivas I, Castro-Díez P, Saldaña A (2015) Analysis of the riparian habitat invasion by three exotic species in Spain. Ecosistemas 24:18–28
Cogolludo-Agustín MÁ, Agúndez D, Gil L (2000) Identification of native and hybrid elms in Spain using isozyme gene markers. Heredity 85:157–166
Collada C, Fuentes-Utrilla P, Gil L, Cervera MT (2004) Characterization of microsatellite loci in Ulmus minor Miller and cross-amplification in U. glabra Hudson and U. laevis Pall. Mol Ecol Notes 4:731–732
Cozzo D (1968) Concepto forestal de la naturalización de especies exóticas y su ocurrencia en Argentina. Rev For Argent 12:118–124
Culley TM, Hardiman NA (2009) The role of intraspecific hybridization in the evolution of invasiveness: a case study of the ornamental pear tree Pyrus calleryana. Biol Invasions 11:1107–1119
Demaio P, Karlin U, Medina M (2015) Árboles nativos de Argentina. Tomo 1: Centro y Cuyo, 1st edn. Ecoval
Drábková L, Kirschner J, Vlček Č (2002) Comparison of seven DNA extraction and amplification protocols in historical herbarium specimens of Juncaceae. Plant Mol Biol Rep 20:161–175
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Eliades N-G, Eliades DG (2009) Haplotype analysis: software for analysis of haplotypes data. Distributed by the authors, Forest Genetics and Forest Tree Breeding, Georg-August University Goettingen, Germany
Ellstrand NC, Schierenbeck KA (2000) Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci 97:7043–7050
Elowsky CG, Jordon-Thaden IE, Kaul RB (2013) A morphological analysis of a hybrid swarm of native Ulmus rubra Muhl. and introduced U. pumila L. (Ulmaceae) in southeastern Nebraska. Phytoneuron 44:1–23
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred form metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gilbert KJ, Andrew RL, Bock DG, Franklin MT, Kane NC, Moore JS, Moyers BT, Renaut S, Rennison DJ, Veen T et al (2012) Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program Structure. Mol Ecol 21:4925–4930
Hirsch H, Wypior C, von Wehrden H, Wesche K, Renison D, Hensen I (2012) Germination performance of native and non-native Ulmus pumila populations. NeoBiota 15:53–68
Hirsch H, Hensen I, Wesche K, Renison D, Wypior C, Hartmann M, von Wehrden H (2016) Non-native populations of an invasive tree outperform their native conspecifics. AoB Plants 8:plw071
Hovick SM, Whitney KD (2014) Hybridisation is associated with increased fecundity and size in invasive taxa: meta-analytic support for the hybridisation-invasion hypothesis. Ecol Lett 17:1464–1477
Hufbauer RA (2004) Population genetics of invasions: can we link neutral markers to management? Weed Technol 18:1522–1527
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Kartesz JT, The Biota of North America Program (BONAP; 2015) North American Plant Atlas. http://www.bonap.org/MapSwitchboard.html. Accessed 07 Dec 2016
Keim P, Paige KN, Whitham TG, Lark KG (1989) Genetic analysis of an interspecific hybrid swarm of Populus: occurrence of unidirectional introgression. Genetics 123:557–565
Kolbe JJ, Glor RE, Schettino LR, Lara AC, Larson A, Losos JB (2004) Genetic variation increases during biological invasion by a cuban lizard. Nature 431:177–181
Kosman E, Leonard KJ (2005) Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol Ecol 14:415–424
Kosman E, Leonard KJ (2007) Conceptual analysis of methods applied to assessment of diversity within and distance between populations with asexual or mixed mode of reproduction. New Phytol 174:683–696
Lachmuth S, Durka W, Schurr FM (2010) The making of a rapid plant invader: genetic diversity and differentiation in the native and invaded range of Senecio inaequidens. Mol Ecol 19:3952–3967
Lavergne S, Molofsky J (2007) Increased genetic diversity and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci USA 104:3883–3888
Le Roux J, Wieczorek AM (2009) Molecular systematics and population genetics of biological invasions: towards a better understanding of invasive species management. Ann Appl Biol 154:1–17
Leopold DJ (1980) Chinese and Siberian elms. J Arboric 6:175–179
Martin M (2008) Comparing invasive species to metastatic cancers inspires new insights for modelers. J Natl Cancer Inst 100:88–89
Mazia CN, Chaneton EJ, Ghersa CM, Leon RJC (2001) Limits to tree species invasion in pampean grassland and forest plant communities. Oecologia 128:594–602
Meusel H, Jäger E, Weinert E (1965) Vergleichende Chorologie der zentraleuropäischen Flora. Teil I. Text und Kartenband. Fischer, Jena
Milne RI, Abbott RJ (2000) Origin and evolution of invasive naturalized material of Rhododendron ponticum L. in the British Isles. Mol Ecol 9:541–556
Mittempergher L, Santini A (2004) The history of elm breeding. Investigación agraria. Sistemas y recursos forestales 13:161–177
Moore G (1960) El cultivo de Ulmus pumila ofrece buenas perspectivas en la Argentina. Rev For Argent 4:77–81
Neher EF, Roic LD (1972) Nota sobre la naturalización de especies arbóreas en las Sierras de Córdoba. Rev For Argent 16:123–125
Nei M (1978) Molecular population genetics and evolution. Columbia University Press, New York
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Poduje L (1972) Contribución al conocimiento del cultivo del olmo siberiano (Ulmus pumila var. arborea Litw.) en La Pampa. Rev For Argent 16:137–141
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Provan J, Soranzo N, Wilson NJ, Goldstein DB, Powell W (1999) A low mutation rate for chloroplast microsatellites. Genetics 153:943–947
R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rejmánek M, Richardson DM (2013) Trees and shrubs as invasive alien species—2013 update of the global database. Divers Distrib 19:1093–1094
Richardson DM, Rejmánek M (2011) Trees and shrubs as invasive alien species—a global review. Divers Distrib 17:788–809
Rieseberg IH, Kim S-C, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K (2007) Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129:149–165
Rius M, Darling JA (2014) How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol 29:233–242
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rosenthal DM, Ramakrishnan AP, Cruzan MB (2008) Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol Ecol 17:4657–4669
Schachtel GA, Dinoor A, Herrmann A, Kosman E (2012) Comprehensive evaluation of virulence and resistance data: a new analysis tool. Plant Dis 96:1060–1063
Schierenbeck KA, Ellstrand NC (2009) Hybridization and the evolution of invasiveness in plants and other organisms. Biol Invasions 11:1093–1105
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Todzia CA, Panero JL (1998) A new species of Ulmus (Ulmaceae) from southern Mexico and a synopsis of the species in Mexico. Brittonia 50:343–347
USDA, NRCS (2011) The PLANTS Database. National Plant Data Team, Greensboro. http://plants.usda.gov. Accessed 5 Oct 2011
Webb WE (1948) A report on Ulmus pumila in the Great Plains region of the United States. J For 46:274–278
Weising K, Gardner RC (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Wesche K, Walther D, von Wehrden H, Hensen I (2011) Trees in the desert: reproduction and genetic structure of fragmented Ulmus pumila forests in Mongolian drylands. Flora 206:91–99
Whiteley RE, Black-Samuelsson S, Clapham D (2003) Development of microsatellite markers for the European white elm (Ulmus laevis Pall.) and cross-species amplification within the genus Ulmus. Mol Ecol Res 3:598–600
Whitney KD, Gabler CA (2008) Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Divers Distrib 14:569–580
Williams DA, Overholt WA, Cuda JP, Hughes CR (2005) Chloroplast and microsatellite DNA diversities reveal the introduction history of Brazilian peppertree (Schinus terebinthifolius) in Florida. Mol Ecol 14:3643–3656
Wu ZY, Raven PH, Hong DY (2003) Flora of China. Vol. 5 (Ulmaceae through Basellaceae). Science Press, Beijin, China and Missouri Botanical Garden Press, St. Louis, USA. http://www.efloras.org. Accessed on 20 Nov 2010
Zalapa JE, Brunet J, Guries RP (2008a) Genetic diversity and relationships among Dutch elm disease tolerant Ulmus pumila L. accessions from China. Genome 51:492–500
Zalapa JE, Brunet J, Guries RP (2008b) Isolation and characterization of microsatellite markers for red elm (Ulmus rubra Muhl.) and cross-species amplification with Siberian elm (Ulmus pumila). Mol Ecol Resour 8:109–112
Zalapa JE, Brunet J, Guries RP (2009) Patterns of hybridization and introgression between invasive Ulmus pumila (Ulmaceae) and native U. rubra. Am J Bot 96:1116–1128
Zalapa JE, Brunet J, Guries RP (2010) The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evol Appl 3:157–168
Zalba SM, Villamil CB (2002) Woody plant invasion in relictual grasslands. Biol Invasions 4:55–72
Acknowledgements
Al Schneider, S. Hegi, R. Peterson, R. Sivinski, J. Smith, T. Frates and B. King directed us to populations and provided hospitality during our field trip in the western United States. Ricardo Suarez collected leaf material in Argentina and Ximing Zhang, Zhenying Huang, B. Oyuntsetseg, L. Yakovchenko in Asia. Denny Walther provided samples from his former research and P. Gebauer, K. Reichel, V. Debus and U. Pietzarka provided samples of U. minor. We also thank A. Suchorukow for providing the sample from the Moscow State University herbarium. Birgit Müller, C. Wypior and E. Gustin contributed to the microsatellite lab work.
Funding
This work was funded by the “Graduiertenförderung des Landes Sachsen-Anhalts” and the DAAD to H. Hirsch, the National Science Foundation Minority Postdoctoral Fellowship to J.E. Zalapa (NSF award #0409651), the USDA-ARS to J. Brunet and the German Academic Exchange Service to M. Hartmann.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hirsch, H., Brunet, J., Zalapa, J.E. et al. Intra- and interspecific hybridization in invasive Siberian elm. Biol Invasions 19, 1889–1904 (2017). https://doi.org/10.1007/s10530-017-1404-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-017-1404-6