Abstract
Functional characterization of metagenomic DNA often involves expressing heterologous DNA in genetically tractable microorganisms such as Escherichia coli. Functional expression of heterologous genes can suffer from limitations due to the lack of recognition of foreign promoters or presence of intrinsic terminators on foreign DNA between a vector-based promoter and the transcription start site. Anti-terminator proteins are a possible solution to overcome this limitation. When bacteriophage lambda infects E. coli, it relies on the host transcription machinery to transcribe and express phage DNA. Lambda anti-terminator protein Q (λQ) regulates the expression of late-genes of phage lambda. E. coli RNA polymerase recognizes the PR' promoter on the lambda genome and forms a complex with λQ, to overcome the terminator tR'. Here we show the use of λQ to efficiently transcribe a capsular polysaccharide cluster, cps3, from Lactobacillus plantarum containing intrinsic terminators in Escherichia coli. In addition, we expand the use of anti-terminator λQ in Pseudomonas putida. The results show ~ fivefold higher expression of a fluorescent reporter located ~ 12.5kbp downstream from the promoter, when the transcription is driven by PR' promoter in presence of λQ compared to a lac promoter. These results suggest that λQ could be used in metabolic engineering to enhance expression of heterologous DNA.



Similar content being viewed by others
References
Calero P, Nikel PI (2019) Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms. Microb Biotechnol 12(1):98–124
Daniels DL, Blattner FR (1982) Nucleotide sequence of the Q gene and the Q to S intergenic region of bacteriophage lambda. Virology 117(1):81–92
Deighan P, Hochschild A (2007) The bacteriophage lambdaQ anti-terminator protein regulates late gene expression as a stable component of the transcription elongation complex. Mol Microbiol 63(3):911–920
Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE (2008) The bacteriophage lambda Q antiterminator protein contacts the beta-flap domain of RNA polymerase. Proc Natl Acad Sci USA 105(40):15305–15310
Elmore JR, Furches A, Wolff GN, Gorday K, Guss AM (2017) Development of a high efficiency integration system and promoter library for rapid modification of Pseudomonas putida KT2440. Metab Eng Commun 5:1–8
Forsberg KJ, Patel S, Witt E, Wang B, Ellison TD, Dantas G (2016) Identification of genes conferring tolerance to lignocellulose-derived inhibitors by functional selections in soil metagenomes. Appl Environ Microbiol 82(2):528–537
Gabor EM, Alkema WB, Janssen DB (2004) Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 6(9):879–886
Ghosh B, Grzadzielska E, Bhattacharya P, Peralta E, DeVito J, Das A (1991) Specificity of antitermination mechanisms. Suppression of the terminator cluster T1–T2 of Escherichia coli ribosomal RNA operon, rrnB, by phage lambda antiterminators. J Mol Biol 222(1):59–66
Grayhack E, Yang X, Lau L, Roberts J (1985) Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell 42(1):259–269
Haynes AR (2014) Characterization of extremophilic bacteria for potential in the biofuel and bioprocess industries. PhD in Plant Pathology and Microbiology, Texas A&M University
Meijnen JP, de Winde JH, Ruijssenaars HJ (2008) Engineering Pseudomonas putida S12 for efficient utilization of D-xylose and L-arabinose. Appl Environ Microbiol 74(16):5031–5037
Mitra A, Angamuthu K, Jayashree HV, Nagaraja V (2009) Occurrence, divergence and evolution of intrinsic terminators across eubacteria. Genomics 94(2):110–116
Nickels BE, Roberts CW, Roberts JW, Hochschild A (2006) RNA-mediated destabilization of the sigma(70) region 4/beta flap interaction facilitates engagement of RNA polymerase by the Q antiterminator. Mol Cell 24(3):457–468
Nikel PI, de Lorenzo V (2018) Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab Eng 50:142–155
Nikel PI, Martinez-Garcia E, de Lorenzo V (2014) Biotechnological domestication of pseudomonads using synthetic biology. Nat Rev Microbiol 12(5):368–379
Peng Q, Wang X, Shang M, Huang J, Guan G, Li Y, Shi B (2014) Isolation of a novel alkaline-stable lipase from a metagenomic library and its specific application for milkfat flavor production. Microb Cell Fact 13:1
Santangelo TJ, Artsimovitch I (2011) Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol 9(5):319–329
Vorobiev SM, Gensler Y, Vahedian-Movahed H, Seetharaman J, Su M, Huang JY, Xiao R, Kornhaber G, Montelione GT, Tong L, Ebright RH, Nickels BE (2014) Structure of the DNA-binding and RNA-polymerase-binding region of transcription antitermination factor lambdaQ. Structure 22(3):488–495
Warren RL, Freeman JD, Levesque RC, Smailus DE, Flibotte S, Holt RA (2008) Transcription of foreign DNA in Escherichia coli. Genome Res 18(11):1798–1805
Acknowledgements
We thank Dr. Michael Benedik for helpful discussion on the use of anti-terminator λQ.
Supplemarntary informations
Supplementary Figure 1: E. coli rpoB homologs in other eubacteria. a An amino acid sequence alignment of the rpoB homologs from various genera highlighting the conserved flap-tip portion (red box) of the β-subunit where λQ is predicted to bind to the RNAP. b The amino acid similarity between E.coli rpoB sequence and the homologs in other genus, including accession numbers for the protein sequences used.
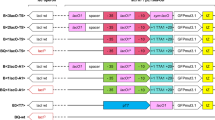
Supplementary Figure 2: Illustration showing insertion of the gene encoding λQ into the P. putida genome. The genes PP_3358-PP_3357-PP_3356 (coding for ech, vdh and fcs respectively) on P. putida chromosome are replaced by the gene encoding λQ with an RBS upstream of it using suicide vector pK18mobsacB.
Supplementary Figure 3: P. putida constructs used in the study. a Genome location showing presence of BxB1-attB site (yellow). b Insertion of the mKate2 (red) - KanR (purple) - Bxb1-attP (yellow) fragment (attP fragment) using phage integrase system. mKate2 does not carry a promoter upstream. c The heterologous DNA (black) with Plac inserted along with the attP fragment on the genome. d The heterologous DNA (black) with PR' inserted along with the attP fragment on the genome in absence of λQ. e The heterologous DNA (black) with PR' inserted along with the attP fragment on the genome in presence of λQ.
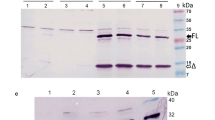
Supplementary Figure 4: No significant growth difference was observed between P. putida strains under study. The strains under study were cultured in M9 minimal media + Kanamycin for ~25 hours in TECAN plate reader. Every 15 minutes, absorbance was measured at 600nm (a) and fluorescence was measured using 588nm/633nm excitation/emission filters (b). All strains show similar growth but varied fluorescence. The growth curve represents an average of triplicates of 3 biological replicates. The error bars represent standard deviation.
Supplementary Table 1: List of strains used in this study.
Supplementary Table 2: List of plasmids used in this study.
Supplementary Table 3: List of promoter and gene sequences used in this study
Supplementary Table 4: Sequences of all the primers used in this study
Funding
This research was supported by Texas A&M President’s Excellence T3 X-grant. This work was in part supported by the Center for Bioenergy Innovation, U.S. DOE Bioenergy Research Center, supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Author information
Authors and Affiliations
Contributions
KCK proposed, funded, and contributed to the analysis of the project. The Pseudomonas putida portion of the paper was funded and supervised by AMG. JAK performed the experiments, collected the data, prepared the figures, and wrote the paper with input from KCK and AMG All authors edited and approved the final version of the paper.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khan, J.A., Guss, A.M. & Kao, K.C. Enhancing transcription in Escherichia coli and Pseudomonas putida using bacteriophage lambda anti-terminator protein Q. Biotechnol Lett 44, 253–258 (2022). https://doi.org/10.1007/s10529-021-03206-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-021-03206-x