Abstract
Objectives
To build a stronger Pichia pastoris PCAT1 promoter and to identify putative transcriptional factor binding sites (TFBSs) on PCAT1 that affect the activity of the promoter.
Result
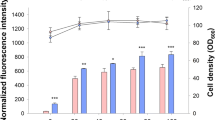
A synthetic library of PCAT1 was generated by deleting or duplicating putative TFBS motifs in the promoter sequence. CSRE, MIG1, RAP1 and HAP2/3/4 were found to have important effects on PCAT1 activity. The PCAT1 variant P4 with a putative binding site of RAP1 on the promoter sequence showed a stronger activity compared with that of the wild-type PCAT1 and PAOX1, which is the strongest natural P. pastoris promoter that has been reported. This inference was confirmed with EGFP (enhanced green fluorescent protein) and Candida Antarctica lipase B as the reporters.
Conclusion
The role of the transcriptional regulator RAP1 may be important in PCAT1 methanol induction. A stronger PCAT1 variant can be constructed by the duplication of the putative binding site of RAP1 on the PCAT1 promoter sequence. This PCAT1 variant has potential value for heterologous protein production, metabolic engineering, and synthetic biology.



Similar content being viewed by others
References
Ata O, Prielhofer R, Gasser B, Mattanovich D, Calik P (2017) Transcriptional engineering of the glyceraldehyde-3-phosphate dehydrogenase promoter for improved heterologous protein production in Pichia pastoris. Biotechnol Bioeng 114:2319–2327. https://doi.org/10.1002/bit.26363
Bi X, Broach JR (1999) UAS(rpg) can function as a heterochromatin boundary element in yeast. Genes Dev 13:1089–1101. https://doi.org/10.1101/gad.13.9.1089
Cartharius K, Frech K, Grote K et al (2005) Matlnspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942. https://doi.org/10.1093/bioinformatics/bti473
Cox RS, Surette MG, Elowitz MB (2007) Programming gene expression with combinatorial promoters. Mol Syst Biol 3:145. https://doi.org/10.1038/msb4100187
Gancedo JM (1998) Yeast carbon catabolite repression. Microbiol Mol Biol Rev 62:334–361. https://doi.org/10.1128/mmbr.62.2.334-361.1998
Gellissen G (2000) Heterologous protein production in methylotrophic yeasts. Appl Microbiol Biotechnol 54:741–750. https://doi.org/10.1007/s002530000464
Hartner FS, Ruth C, Langenegger D et al (2008) Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res 36:76. https://doi.org/10.1093/nar/gkn369
Jayaram N, Usvyat D, Martin ACR (2016) Evaluating tools for transcription factor binding site prediction. BMC Bioinform 17:547. https://doi.org/10.1186/s12859-016-1298-9
Juturu V, Wu JC (2018) Heterologous protein expression in Pichia pastoris: latest research progress and applications. ChemBioChem 19:7–21. https://doi.org/10.1002/cbic.201700460
Prielhofer R, Reichinger M, Wagner N et al (2018) Superior protein titers in half the fermentation time: promoter and process engineering for the glucose-regulated GTH1 promoter of Pichia pastoris. Biotechnol Bioeng 115:2479–2488. https://doi.org/10.1002/bit.26800
Roth S, Kumme J, Schuller HJ (2004) Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr Genet 45:121–128. https://doi.org/10.1007/s00294-003-0476-2
Vogl T, Sturmberger L, Kickenweiz T et al (2016) A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth Biol 5:172–186. https://doi.org/10.1021/acssynbio.5b00199
Xu N, Wei L, Liu J (2019) Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World J Microbiol Biotechnol 35:33. https://doi.org/10.1007/s11274-019-2606-0
Zhang XW, Zhang XQ, Liang SL, Ye YR, Lin Y (2013) Key regulatory elements of a strong constitutive promoter, P-GCW14, from Pichia pastoris. Biotechnol Lett 35:2113–2119. https://doi.org/10.1007/s10529-013-1312-5
Zhu TC, Sun HB, Wang MY, Li Y (2019) Pichia pastoris as a versatile cell factory for the production of industrial enzymes and chemicals: current status and future perspectives. Biotechnol J 14:1800694. https://doi.org/10.1002/biot.201800694
Acknowledgements
This work was supported by Guangdong Key Area Research and Development Program (Grant No. NO. 2019B020210003).
Supplementary information
Supplementary Figure 1−Plasmid used in this study.
Supplementary Table S1−Primers used in this study.
Supplementary Table S2−The putative TFBSs searching result on PCAT1 by MatInspector.
Supplementary Table S3−Frequency of putative TFBSs matrix family occurrence within 16 kinds of methanol-inducible promoters.
Supplementary Table S4−Frequency of putative TFBSs matrix occurrence within 16 kinds of methanol-inducible promoters.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nong, L., Zhang, Y., Duan, Y. et al. Engineering the regulatory site of the catalase promoter for improved heterologous protein production in Pichia pastoris. Biotechnol Lett 42, 2703–2709 (2020). https://doi.org/10.1007/s10529-020-02979-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-02979-x