Abstract
Objectives
Developing a dynamic regulation strategy is an essential step in establishing an automatic control system for manipulating metabolic fluxes and cellular behaviors. To broaden the extent of the application, a system that can generally control any gene of interest is demanded.
Results
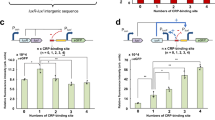
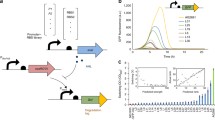
Through characterization and optimization, the strategy repressed the immediate expression incrementally from 0 to 90% during culturing. Moreover, by changing single base pair in the lux box of the Plux promoter, the degree of repression of the target genomic gene was tuned to a difference of 70%. This strategy is expected to control metabolic flux without disrupting cell growth.
Conclusions
We engineered bacterial small RNA to develop a pathway-independent strategy that can dynamically repress the expression of any gene at the posttranscription level.




Similar content being viewed by others
Abbreviations
- E. coli :
-
Escherichia coli
- AHL:
-
3-Oxohexanoylhomoserine lactone
- sRNA:
-
Small RNA
- sfGFP:
-
Super-folded green fluorescent protein
- TBS:
-
Target binding sequence
- QS:
-
Quorum sensing
- OPPS:
-
Octaprenyl pyrophosphate synthase
- GOI:
-
Gene of interest
References
Asai K, Fujisaki S, Nishimura Y, Nishino T, Okada K, Nakagawa T, Kawamukai M, Matsuda H (1994) The identification of Escherichia coli ispB (cel) gene encoding the octaprenyl diphosphate synthase. Biochem Biophys Res Commun 202:340–345. https://doi.org/10.1006/bbrc.1994.1933
Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L (2008) A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol 4:187. https://doi.org/10.1038/msb.2008.24
Cataldi TR, Bianco G, Palazzo L, Quaranta V (2007) Occurrence of N-acyl-L-homoserine lactones in extracts of some Gram-negative bacteria evaluated by gas chromatography-mass spectrometry. Anal Biochem 361:226–235. https://doi.org/10.1016/j.ab.2006.11.037
Gupta A, Reizman IM, Reisch CR, Prather KL (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35:273–279. https://doi.org/10.1038/nbt.3796
Kim EM, Woo HM, Tian T, Yilmaz S, Javidpour P, Keasling JD, Lee TS (2017) Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab Eng 44:325–336. https://doi.org/10.1016/j.ymben.2017.11.004
Kimura Y, Tashiro Y, Saito K, Kawai-Noma S, Umeno D (2016) Directed evolution of Vibrio fischeri LuxR signal sensitivity. J Biosci Bioeng 122:533–538. https://doi.org/10.1016/j.jbiosc.2016.04.010
Liu CJ, Jiang H, Wu L, Zhu LY, Meng E, Zhang DY (2017) OEPR Cloning: an efficient and seamless cloning strategy for large- and multi-fragments. Sci Rep 7:44648. https://doi.org/10.1038/srep44648
Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol (Clifton, NJ) 453:3–31. https://doi.org/10.1007/978-1-60327-429-6_1
Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J (2011) A sensing array of radically coupled genetic ‘biopixels’. Nature 481:39–44. https://doi.org/10.1038/nature10722
Scott SR, Hasty J (2016) Quorum sensing communication modules for microbial consortia. ACS Synth Biol 5:969–977. https://doi.org/10.1021/acssynbio.5b00286
Soma Y, Hanai T (2015) Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng 30:7–15. https://doi.org/10.1016/j.ymben.2015.04.005
Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, Fields S, Church GM, Raman S (2016) Engineering an allosteric transcription factor to respond to new ligands. Nat Methods 13:177–183. https://doi.org/10.1038/nmeth.3696
You L, Cox RS 3rd, Weiss R, Arnold FH (2004) Programmed population control by cell-cell communication and regulated killing. Nature 428:868–871. https://doi.org/10.1038/nature02491
Zeng W, Du P, Lou Q, Wu L, Zhang HM, Lou C, Wang H, Ouyang Q (2017) Rational design of an ultrasensitive quorum-sensing switch. ACS Synth Biol 6:1445–1452. https://doi.org/10.1021/acssynbio.6b00367
Zhang F, Carothers JM, Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 30:354–359. https://doi.org/10.1038/nbt.2149
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant Number 81703400 (to Dr. E. Meng). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
EM, D-YZ and S-HB designed the appropriate technology routes. S-HB conducted main experiments including plasmid and strain construction, shake flask fermentation and western blot experiments. EM, W-YL, C-JL and D-YZ participated in the establishment of mathematic models. EM and D-YZ supervised the project and S-HB wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bao, SH., Li, WY., Liu, CJ. et al. Quorum-sensing based small RNA regulation for dynamic and tuneable gene expression. Biotechnol Lett 41, 1147–1154 (2019). https://doi.org/10.1007/s10529-019-02719-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02719-w