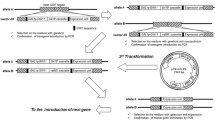
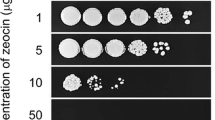
Abstract
Objectives
To establish a recombinase flippase (FLP) and flippase recognition target (FRT) system-mediated protocol for post-integration excision of exogenous DNA fragments in the oleaginous yeast Rhodosporidium toruloides.
Results
Binary vectors were constructed to harbor FLP expressing cassette together with the hygromycin-resistance marker. Results showed that R. toruloides transformants produced FLP, but failed to mediate removal of the bleomycin-resistance marker within two FRT sites. When FLP was fused with a native nuclear localization signal (NLS) peptide, the system was found functional. Moreover, R. toruloides recombinant strains expressing the NLS-fused FLP under the control of PADH2, an promoter of alcohol dehydrogenase 2 gene (RHTO_03062), were obtained to realize homologous recombination upon growing in glucose-deficient medium.
Conclusions
We have devised a homologous recombination method for R. toruloides based on the FLP/FRT system, which may facilitate further metabolic engineering and designing advanced cell factories for value-added chemicals.



Similar content being viewed by others
References
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Hu C, Zhao X, Zhao J, Wu S, Zhao ZK (2009) Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour Technol 100:4843–4847
Koh CMJ, Liu Y, Moehninsi D, Du M, Ji L (2014) Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol 14:50
Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH (2007) Classical nuclear localization signals: definition, function, and interaction with import in alpha. J Biol Chem 282:5101–5105
Lin X, Wang Y, Zhang S, Zhu Z, Zhou YJ, Yang F, Sun W, Wang X, Zhao ZK (2014) Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res 14:547–555
Liu Y, Koh CMJ, Sun L, Hlaing M, Du M, Peng N, Ji L (2013) Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides. Appl Microbiol Biotechnol 97:719–729
Liu H, Jiao X, Wang Y, Yang X, Sun W, Wang J, Zhang S, Zhao ZK (2017) Fast and efficient genetic transformation of oleaginous yeast Rhodosporidium toruloides by using electroporation. FEMS Yeast Res 17:fox017
Long D, Lu W, Hao Z, Xiang Z, Zhao A (2016) Highly efficient and inducible DNA excision in transgenic silkworms using the FLP/FRT site-specific recombination system. Transgenic Res 25:795–811
Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CN Jr, McAvoy R, Jiang X, Wu Y, He A, Pei Y, Li Y (2007) ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5:263–274
Park Y-K, Nicaud J-M, Ledesma-Amaro R (2018) The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol 36:304–317
Schweizer HP (2003) Applications of the Saccharomyces cerevisiae Flp-FRT system in bacterial genetics. J Mol Microb Biotechnol 5:67–77
Tan X, Liang F, Cai K, Lu X (2013) Application of the FLP/FRT recombination system in cyanobacteria for construction of markerless mutants. Appl Microbiol Biotechnol 97:6373–6382
Tsai YY, Ohashi T, Kanazawa T, Polburee P, Misaki R, Limtong S, Fujiyama K (2017) Development of a sufficient and effective procedure for transformation of an oleaginous yeast, Rhodosporidium toruloides DMKU3-TK16. Curr Genet 63:359–371
Van den Ent F, Lowe J (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67:67–74
Wang Y, Zhang S, Pötter M, Sun W, Li L, Yang X, Jiao X, Zhao ZK (2016) Overexpression of delta 12-fatty acid desaturase in the oleaginous yeast Rhodosporidium toruloides for production of linoleic acid-rich lipids. Appl Biochem Biotechnol 180:1497–1507
Yaegashi J, Kirby J, Ito M, Sun J, Dutta T, Mirsiaghi M, Sundstrom ER, Rodriguez A, Baidoo E, Tanjore D, Pray T, Sale K, Singh S, Keasling JD, Simmons BA, Singer SW, Magnuson JK, Arkin AP, Skerker JM, Gladden JM (2017) Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels 10:241
Zhang S, Ito M, Skerker JM, Arkin AP, Rao CV (2016) Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl Microbiol Biotechnol 100:9393–9405
Acknowledgement
This work was supported by National Natural Science Foundation of China (51761145014, 21602218).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, W., Yang, X., Wang, X. et al. Developing a flippase-mediated maker recycling protocol for the oleaginous yeast Rhodosporidium toruloides. Biotechnol Lett 40, 933–940 (2018). https://doi.org/10.1007/s10529-018-2542-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2542-3