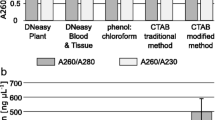
Abstract
Many protocols have been used for extraction of DNA from Thraustochytrids. These generally involve the use of CTAB, phenol/chloroform and ethanol. They also feature mechanical grinding, sonication, N2 freezing or bead beating. However, the resulting chemical and physical damage to extracted DNA reduces its quality. The methods are also unsuitable for large numbers of samples. Commercially-available DNA extraction kits give better quality and yields but are expensive. Therefore, an optimized DNA extraction protocol was developed which is suitable for Thraustochytrids to both minimise expensive and time-consuming steps prior to DNA extraction and also to improve the yield. The most effective method is a combination of single bead in TissueLyser (Qiagen) and Proteinase K. Results were conclusive: both the quality and the yield of extracted DNA were higher than with any other method giving an average yield of 8.5 µg/100 mg biomass.



Similar content being viewed by others
References
Ahmed OB, Asghar AH, Elhassan MM (2014) Comparison of three DNA extraction methods for polymerase chain reaction (PCR) analysis of bacterial genomic DNA. Afr J Microbiol Res 8:598–602
Borman AM, Palmer M, Johnson EM (2012) Rapid methods for the extraction and archiving of molecular grade fungal genomic DNA. Methods Mol Biol 968:55–62
Cavalier-Smith T, Allsopp MTEP, Chao EE (1994) Thraustochytrids are chromists, not fungi: 18S rRNA signatures of Heterokonta. Philos Trans R Soc B 346:387–397
Chang KJL, Dunstan GA, Abell GCJ et al (2012) Biodiscovery of new Australian thraustochytrids for production of biodiesel and long-chain omega-3 oils. Appl Microbiol Biotechnol 93:2215–2231
González-Mendoza D, Argumedo-Delira A, Morales-Trejo A et al (2010) A rapid method for isolation of total DNA from pathogenic filamentous plant fungi. Genet Mol Res 9:162–166
Graham GC, Mayers P, Henry RJ (1994) A simplified method for the preparation of fungal genomic DNA for PCR and RAPD analysis. Biotechniques 16:48–50
Honda D, Yokochi T, Nakahara T et al (1999) Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J Eukaryot Microbiol 46:637–647
Jakobsen AN (2008) Compatible solutes and docosahexaenoic acid accumulation of thraustochytrids of the Aurantiochytrium group. Norwegian University of Science and Technology, Norway
Jakobsen AN, Aasen IM, Josefsen KD, Strom AR (2008) Accumulation of docosahexaenoic acid-rich lipid in thraustochytrid Aurantiochytrium sp strain T66: effects of N and P starvation and O-2 limitation. Appl Microbiol Biotechnol 80:297–306
Kimura H, Fukuba T, Naganuma T (1999) Biomass of thraustochytrid protoctists in coastal water. Mar Ecol Prog Ser 189:27–33
Mo C, Rinkevich B (2001) A simple, reliable, and fast protocol for thraustochytrid DNA extraction. Mar Biotechnol 3:100–102
Moss ST (1986) Biology and phylogeny of the Labyrinthulaes and Thraustochytriales. In: Moss ST (ed) The biology of marine fungi. Cambridge University Press, Cambridge, pp 105–129
Osmundson TW, Eyre CA, Hayden KM, Dhillon J, Garbelotto MM (2013) Back to basics: an evaluation of NaOH and alternative rapid DNA extraction protocols for DNA barcoding, genotyping, and disease diagnostics from fungal and oomycete samples. Mol Ecol Resour 13:66–74
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria URL http://www.R-project.org/
Tendulkar SR, Gupta A, Chattoo BB (2003) A simple protocol for isolation of fungal DNA. Biotechnol Lett 25:1941–1944
Acknowledgments
We would like to thank Dr. Tom Lewis (Tasmania), Solaris Bioscience (Tasmania) and Queensland University of Technology (Queensland) for their support of this project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ranasinghe, C.P., Harding, R. & Hargreaves, M. An improved protocol for the isolation of total genomic DNA from Labyrinthulomycetes. Biotechnol Lett 37, 685–690 (2015). https://doi.org/10.1007/s10529-014-1712-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1712-1