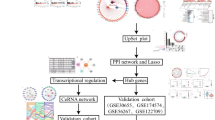
Abstract
Osteoarthritis (OA) is a serious threat to human health. However, the etiology and pathogenesis of the disease are not fully understood. Most researchers believe that the degeneration and imbalance of articular cartilage, extracellular matrix, and subchondral bone are the fundamental causes of osteoarthritis. However, recent studies have shown that synovial lesions may precede cartilage, which may be an important precipitating factor in the early stage of OA and the whole course of the disease. This study aimed to conduct an analysis based on sequence data from the Gene Expression Omnibus (GEO) database to investigate the presence of effective biomarkers in the synovial tissue of osteoarthritis for the diagnosis and control of OA progression. In this study, the differentially expressed OA-related genes (DE-OARGs) in osteoarthritis synovial tissues were extracted in the GSE55235 and GSE55457 datasets using the Weighted Gene Co-expression Network Analysis (WGCNA) and limma. Least-Absolute Shrinkage and Selection Operator (LASSO) algorithm was used to select the diagnostic genes based on the DE-OARGs by glmnet package. 7 genes were selected as diagnostic genes including SAT1, RLF, MAFF, SIK1, RORA, ZNF529, and EBF2. Subsequently, the diagnostic model was constructed and the results of the Area Under the Curve (AUC) demonstrated that the diagnostic model had high diagnostic performance for OA. Additionally, among the 22 immune cells of the Cell type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) and the 24 immune cells of the single sample Gene Set Enrichment Analysis (ssGSEA), 3 immune cells and 5 immune cells were different between the OA and normal samples, respectively. The expression trends of the 7 diagnostic genes were consistent in the GEO datasets and the results of the real-time reverse transcription PCR (qRT-PCR). The results of this study demonstrate that these diagnostic markers have important significance in the diagnosis and treatment of OA, and will provide further evidence for the clinical and functional studies of OA.









Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- RF:
-
Random forest
- DT:
-
Decision tree
- SVM:
-
Support vector machine
- MM:
-
Module membership
- GS:
-
Gene significance
- LASSO:
-
Least-Absolute Shrinkage and Selection Operator
- OA:
-
Osteoarthritis
- GEO:
-
Gene Expression Omnibus
- WGCNA:
-
Weighted Gene Co-expression Network Analysis
- DE-OARGs:
-
Differentially expressed OA-related genes
- OARGs:
-
OA-related genes
- AUC:
-
Area Under the Curve
- ROC:
-
Receiver operating characteristic
- CIBERSORT:
-
Cell type Identification By Estimating Relative Subsets Of RNA Transcripts
- ssGSEA:
-
Gene Set Enrichment Analysis
- PRP:
-
Platelet-rich plasma
- GO:
-
Gene Ontology
- KEGG:
-
The Kyoto Encyclopedia of Genes and Genomes
- GSEA:
-
Gene set enrichment analysis
- DGIdb:
-
Drug-Gene Interaction database
- qRT-PCR:
-
Real-time reverse transcription PCR
- DEGs:
-
Differentially expressed genes
- PCA:
-
Principal component analysis
- FC:
-
Fold change
References
Alvarez-Garcia O et al (2016) Increased DNA methylation and reduced expression of transcription factors in human osteoarthritis cartilage. Arthritis Rheumatol 68(8):1876–1886
Bratus-neuenschwander A et al (2018) Pain-associated transcriptome changes in synovium of knee osteoarthritis patients. Gene 9(7):338
Cao J et al (2021) Weighted gene co-expression network analysis reveals specific modules and hub genes related to immune infiltration of osteoarthritis. Ann Transl Med 9(20):1525
Chen B et al (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259
Delanois RE et al (2022) Biologic therapies for the treatment of knee osteoarthritis: an updated systematic review. J Arthroplasty S0883–5403(22):00582–00584
Ellam J, Berenbaum F (2010) The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol 6(11):625–635
Friedman J, Hastie T, Tibshirani R (2010) Regularization Paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22
Grieshaber-Bouyer R et al (2019) Divergent mononuclear cell participation and cytokine release profiles define hip and knee osteoarthritis. J Clin Med 8(10):1631
Guangchuang Yu (2021). enrichplot: Visualization of Functional Enrichment Result. R packageversion 1.10.2 https://yulab-smu.top/biomedical-knowledge-mining-book/.
Guermazi A et al (2011) Assessment of synovitis with contrast-enhanced MRI using a whole-joint semiquantitative scoring system in people with, or at high risk of, knee osteoarthritis: the MOST study. Ann Rheum Dis 70(5):805–811
Keen HI et al (2015) Ultrasound assessment of response to intra-articular therapy in osteoarthritis of the knee. Rheumatology 54(8):1385–1391
Kieslinger M et al (2005) EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev Cell 9(6):757–767
Kim MK et al (2019) Salt-inducible kinase 1 regulates bone anabolism via the CRTC1-CREB-Id1 axis. Cell Death Dis 10(11):826
Kishiro S et al (2008) Selinidin suppresses IgE-mediated mast cell activation by inhibiting multiple steps of Fc epsilonRI signaling. Biol Pharm Bull 31(3):442–448
Jeffrey T. Leek, W. Evan Johnson, Hilary S. Parker, Elana J. Fertig, Andrew E. Jaffe, Yuqing Zhang, John D. Storey and Leonardo Collado Torres (2020). sva: Surrogate Variable Analysis. R package version 3.38.0.
Li MH et al (2017) Regenerative approaches for cartilage repair in the treatment of osteoarthritis. Osteoarthritis Cartilage 25(10):1577–1587
Li J, Yang X, Chu Q, Xie L, Ding Y, Xu X, Timko MP, Fan L (2022) Multi-omics molecular biomarkers and database of osteoarthritis. Database (Oxford) 2022:baac052
Li X et al (2022) Transcriptome analysis reveals candidate lignin-related genes and transcription factors during fruit development in pomelo (Citrus maxima). Genes (basel) 13(5):845
Liang T et al (2021) Inhibition of nuclear receptor RORα attenuates cartilage damage in osteoarthritis by modulating IL-6/STAT3 pathway. Cell Death Dis 12(10):886
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2(3):18–22
Mimpen JY et al (2021) Interleukin-17A causes osteoarthritis-like transcriptional changes in human osteoarthritis-derived chondrocytes and synovial fibroblasts in vitro. Front Immunol 12(12):676173
Nees TA et al (2020) T Helper cell infiltration in osteoarthritis-related knee pain and disability. J Clin Med 9(8):2423
Rego-Pérez I, Durán-Sotuela A, Ramos-Louro P, Blanco FJ (2021) Genetic biomarkers in osteoarthritis: a quick overview. Fac Rev 10(10):78
Rim YA, Ju JH (2020) The Role of fibrosis in osteoarthritis progression. Life (basel) 11(1):3
Ritchie ME et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47
Robin X et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 17(12):77
Scanzello CR et al (2014) Synovial inflammation in patients undergoing arthroscopic meniscectomy: molecular characterization and relationship to symptoms. Arthritis Rheum 63(2):391–400
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Tavallaee G et al (2020) MicroRNAs in synovial pathology associated with osteoarthritis. Front Med (lausanne) 11(7):376
Thomson A, Hilkens CMU (2021) Synovial macrophages in osteoarthritis: the key to understanding pathogenesis? Front Immunol 15(12):678757
Wang G et al (2021) Neutrophil Elastase induces chondrocyte apoptosis and facilitates the occurrence of osteoarthritis via caspase signaling pathway. Front Pharmacol 14(12):666162
Wickham H (2016) Elegant Graphics for Data Analysis. Springer-Verlag, NewYork
Woods A et al (2009) Control of chondrocyte gene expression by actin dynamics: a novel role of cholesterol/Ror-alpha signalling in endochondral bone growth. J Cell Mol Med 13(9B):3497–3516
Yu G et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287
Zhang H et al (2022) CREB ameliorates osteoarthritis progression through regulating chondrocytes autophagy via the miR-373/METTL3/TFEB Axis. Front Cell Dev Biol 9(9):778941
Zhao H, Gong N (2019) miR-20a regulates inflammatory in osteoarthritis by targeting the IκBβ and regulates NK-κB signaling pathway activation. Biochem Biophys Res Commun 518(4):632–637
Funding
No.
Author information
Authors and Affiliations
Contributions
Y-SZ and HY are co-first authors. Y-SZ, HY, and CJ contributed to the conception and design of the research. T-TM and J-NZ performed the experiments. Y-SZ, HY, and CJ analyzed data, interpreted the results of the experiments, wrote the main manuscript text. T-TM and J-NZ prepared all the figures and the table. All authors reviewed the manuscript and approved the final version of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Approval
The qRT-PCR experiments of synovial tissues in our study were promised by the the Ethics Committee of the First People’s Hospital of Wenling (KY-2022–1007-01).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, YS., Yan, H., Mo, TT. et al. Identification of Diagnostic Markers in Synovial Tissue of Osteoarthritis by Weighted Gene Coexpression Network. Biochem Genet 61, 2056–2075 (2023). https://doi.org/10.1007/s10528-023-10359-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10359-z