Abstract
Lung adenocarcinoma (LUAD) is an aggressive malignancy with a poor prognosis. In this study, we explored the critical role and mechanism of circ_0010235 in the pathogenesis of LUAD. The expression levels of circ_0010235, microRNA (miR)-1249-3p, and homeobox A13 (HOXA13) were gauged by quantitative real-time PCR (qRT-PCR) and Western blot. Cell proliferation, cycle progression, migration, and invasion were evaluated by Cell Counting Kit-8 (CCK-8), 5-Ethynyl-2′-Deoxyuridine (Edu), flow cytometry, and transwell assays, respectively. The direct relationship between miR-1249-3p and circ_0010235 or HOXA13 was validated by dual-luciferase reporter, RNA immunoprecipitation (RIP), and RNA pull-down assays. Xenograft experiments were used to examine the role of circ_0010235 in vivo. Circ_0010235 was significantly overexpressed in human LUAD. Silencing of circ_0010235 hindered LUAD cell proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT) in vitro, as well as diminished tumor growth in vivo. Mechanistically, circ_0010235 targeted and inhibited miR-1249-3p. Moreover, circ_0010235 depletion repressed cell malignant behaviors by upregulating miR-1249-3p. HOXA13 was identified as a direct and functional target of miR-1249-3p. Furthermore, circ_0010235 regulated HOXA13 expression by competing for shared miR-1249-3p. Our findings demonstrate that the circ_0010235/miR-1249-3p/HOXA13 axis is implicated in the pathogenesis of LUAD.









Similar content being viewed by others
Data Availability
Please contact the correspondence author for the data request.
References
Abdel Ghafar MT, Elkhouly RA, Elnaggar MH, Mabrouk MM, Darwish SA, Younis RL et al (2021) Utility of serum neuropilin-1 and angiopoietin-2 as markers of hepatocellular carcinoma. J Investig Med 69:1222–1229
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
Deng Y, He R, Zhang R, Gan B, Zhang Y, Chen G et al (2018) The expression of HOXA13 in lung adenocarcinoma and its clinical significance: a study based on the cancer genome Atlas, oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol Lett 15:8556–8572
Denisenko TV, Budkevich IN, Zhivotovsky B (2018) Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 9:117
Ding J, Wu W, Yang J, Wu M (2019) Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249–3p/HOXB8 axis. Pathol Res Pract 215:152376
Dong Y, Cai Y, Liu B, Jiao X, Li ZT, Guo DY et al (2017) HOXA13 is associated with unfavorable survival and acts as a novel oncogene in prostate carcinoma. Future Oncol 13:1505–1516
El-Guindy DM, Wasfy RE, Abdel Ghafar MT, Ali DA, Elkady AM (2019) Oct4 expression in gastric carcinoma: association with tumor proliferation, angiogenesis and survival. J Egypt Natl Canc Inst 31:3
Ghafar MTA, Gharib F, Al-Ashmawy GM, Mariah RA (2020) Serum high-temperature-required protein A2: a potential biomarker for the diagnosis of breast cancer. Gene Rep 20:100706
Gu ZD, Shen LY, Wang H, Chen XM, Li Y, Ning T et al (2009) HOXA13 promotes cancer cell growth and predicts poor survival of patients with esophageal squamous cell carcinoma. Cancer Res 69:4969–4973
Gu Y, Gu J, Shen K, Zhou H, Hao J, Li F et al (2020) HOXA13 promotes colon cancer progression through β-catenin-dependent WNT pathway. Exp Cell Res 395:112238
Habib EM, Nosiar NA, Eid MA, Taha AM, Sherief DE, Hassan AE et al (2021) MiR-150 Expression in Chronic Myeloid Leukemia: Relation to Imatinib Response. Lab Med. https://doi.org/10.1093/labmed/lmab040
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E et al (2010) Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 18:282–293
Hutchinson BD, Shroff GS, Truong MT, Ko JP (2019) Spectrum of lung adenocarcinoma. Semin Ultrasound CT MR 40:255–264
Iwakawa HO, Tomari Y (2015) The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25:651–665
Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH et al (2018) Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol 144:667–674
Kashiwabara K, Fuji S, Tsumura S, Sakamoto K (2020) Prognosis of EGFR-mutant Lung adenocarcinoma patients with malignant pleural effusion receiving first-line EGFR-TKI therapy without Pleurodesis: a single-institute retrospective study. Anticancer Res 40:1117–1121
Kristensen LS, Hansen TB, Venø MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37:555–565
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691
Kumar S, Sharawat SK, Ali A, Gaur V, Malik PS, Pandey M et al (2020) Differential expression of circulating serum miR-1249-3p, miR-3195, and miR-3692-3p in non-small cell lung cancer. Hum Cell 33:839–849
Liang L, Zhang L, Zhang J, Bai S, Fu H (2020) Identification of circRNA-miRNA-mRNA networks for exploring the fundamental mechanism in lung adenocarcinoma. Onco Targets Ther 13:2945–2955
Ma D, Liu H, Qin Y, Li D, Cui Y, Li L et al (2020) Circ_0007142/miR-186/FOXK1 axis promoted lung adenocarcinoma progression. Am J Transl Res 12:4728–4738
Ma L, Zhang X, Yu K, Xu X, Chen T, Shi Y et al (2021) Targeting SLC3A2 subunit of system X(C)(-) is essential for m(6)A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic Biol Med 168:25–43
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Meza R, Meernik C, Jeon J, Cote ML (2015) Lung cancer incidence trends by gender, race and histology in the United States, 1973–2010. PLoS ONE 10:e0121323
Shu H, Hu J, Deng H (2019) miR-1249–3p accelerates the malignancy phenotype of hepatocellular carcinoma by directly targeting HNRNPK. Mol Genet Genomic Med 7:e00867
Siriwardhana C, Khadka VS, Chen JJ, Deng Y (2019) Development of a miRNA-seq based prognostic signature in lung adenocarcinoma. BMC Cancer 19:34
Suster DI, Mino-Kenudson M (2020) Molecular pathology of primary non-small cell lung cancer. Arch Med Res 51:784–798
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014; 505:344–352.
Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu Y et al (2019) RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer 18:134
Wang Y, He B, Dong Y, He GJ, Qi XW, Li Y et al (2021) Homeobox-A13 acts as a functional prognostic and diagnostic biomarker via regulating P53 and Wnt signaling pathways in lung cancer. Cancer Biomark. https://doi.org/10.3233/CBM-200540
Wen Y, Shu F, Chen Y, Chen Y, Lan Y, Duan X et al (2018) The prognostic value of HOXA13 in solid tumors: a meta-analysis. Clin Chim Acta 483:64–68
Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM et al (2011) Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473:101–104
Xin T, Li S, Zhang Y, Kamali X, Liu H, Jia T (2020) circRNA Hsa_circ_0020850 silence represses the development of lung adenocarcinoma via regulating miR-195-5p/IRS2 Axis. Cancer Manag Res 12:10679–10692
Yao Y, Hua Q, Zhou Y (2019) CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun 508:37–45
Yerukala Sathipati S, Ho SY (2017) Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep 7:7507
Yin N, Liu Y, Khoor A, Wang X, Thompson EA, Leitges M et al (2019) Protein kinase Cι and Wnt/β-catenin signaling: alternative pathways to Kras/Trp53-driven lung adenocarcinoma. Cancer Cell 36:156-167.e157
Yu N, Yong S, Kim HK, Choi YL, Jung Y, Kim D et al (2019) Identification of tumor suppressor miRNAs by integrative miRNA and mRNA sequencing of matched tumor-normal samples in lung adenocarcinoma. Mol Oncol 13:1356–1368
Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YN, Zhang YJ (2020) CircRNA EPB41L2 inhibits tumorigenicity of lung adenocarcinoma through regulating CDH4 by miR-211-5p. Eur Rev Med Pharmacol Sci 24:3749–3760
Zhang F, Cheng R, Li P, Lu C, Zhang G (2021) Hsa_circ_0010235 functions as an oncogenic drive in non-small cell lung cancer by modulating miR-433-3p/TIPRL axis. Cancer Cell Int 21:73
Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z (2019) CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis 10:885
Zhu Y, Ma C, Lv A, Kou C (2021) Circular RNA circ_0010235 sponges miR-338-3p to play oncogenic role in proliferation, migration and invasion of non-small-cell lung cancer cells through modulating KIF2A. Ann Med 53:693–706
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
JL was responsible for drafting the manuscript. JL and ZF contributed to the analysis and interpretation of the data. JL and ZF contributed in the data collection. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no financial conflict of interest.
Ethical Approval
Written informed consent was obtained from patients with approval by the Institutional Review Board in Shanxi Provincial Cancer Hospital.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2021_10181_MOESM2_ESM.tif
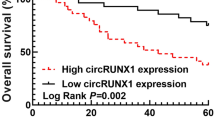
Supplementary file2 (TIF 354 KB) Supplement Figure 1 Correlation between the expression of circ_0010235, miR-1249-3p or HOXA13 and the overall survival of patients. All patients were divided into two groups according to the median of circ_0010235, miR-1249-3p or HOXA13 expression. A Correlation between circ_0010235 expression and patient overall survival. B Correlation between miR-1249-3p expression and patient overall survival. C Correlation between HOXA13 expression and patient overall survival
10528_2021_10181_MOESM3_ESM.tif
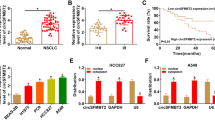
Supplementary file3 (TIF 2991 KB) Supplement Figure 2 Selection of HOXA13 in this study. A Venn diagram showing the putative targets of miR-1249-3p predicted by both Starbase3.0 and TargetScan7.2. B–I Correlation between the expression of eight genes and patient survival analyzed by the Starbase3.0 database. J qRT-PCR of four genes in LUAD tissues and matched nontumor tissues from the same patients (n = 3). *P < 0.05
Rights and permissions
About this article
Cite this article
Liu, J., Fu, Z. Identification of a Novel circ_0010235/miR-1249-3p/HOXA13 Axis in Lung Adenocarcinoma. Biochem Genet 60, 1657–1675 (2022). https://doi.org/10.1007/s10528-021-10181-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10181-5