Abstract
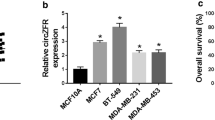
Some circular RNAs (circRNAs) have been verified to act as essential regulators in the progression of breast cancer (BC). We aimed to investigate the role of circRNA trefoil factor 1 (circ-TFF1) in BC progression. The expression of circ-TFF1, microRNA-338-3p (miR-338-3p) and fibroblast growth factor receptor 1 (FGFR1) mRNA was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation was evaluated by methylthiazolyldiphenyl-tetrazolium bromide (MTT), colony formation, and 5-Ethynyl-2′-deoxyuridine (EDU) assays. Cell apoptosis and invasion were assessed by flow cytometry and transwell assay, respectively. Cellular glycolysis, including glucose consumption, lactate production, and ATP/ADP ratio, was detected by commercial kits. All protein levels were measured by western blot assay. The relationship between miR-338-3p and circ-TFF1 or FGFR1 was predicted by online bioinformatics tool and verified by dual-luciferase reporter assay. Xenograft tumor model was established to verify the function of circ-TFF1 in vivo. Circ-TFF1 was overexpressed in BC tissues and cells. Circ-TFF1 knockdown inhibited cell proliferation, invasion and glycolysis and induced apoptosis in BC cells. Circ-TFF1 acted as a sponge of miR-338-3p, and the effects of circ-TFF1 knockdown on BC cell proliferation, apoptosis, invasion, and glycolysis were abolished by miR-338-3p inhibition. FGFR1 was confirmed to be a target gene of miR-338-3p, and miR-338-3p played a tumor-suppressive role in BC by targeting FGFR1. Moreover, circ-TFF1 regulated FGFR1 expression by targeting miR-338-3p. Additionally, circ-TFF1 knockdown hampered tumorigenesis in vivo. Circ-TFF1 knockdown suppressed BC progression by regulating miR-338-3p/FGFR1 axis, providing a promising therapeutic target for BC.









Similar content being viewed by others
Data Availability
Please contact the correspondence author for the data request. The data that support the findings of this study are available from the corresponding author.
References
Akram M, Iqbal M, Daniyal M, Khan AU (2017) Awareness and current knowledge of breast cancer. Biol Res 50:33
Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH (2019) CircRNAs and cancer: Biomarkers and master regulators. Semin Cancer Biol 58:90–99
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A Cancer Journal for Clinicians 68:394–424
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388
Duan X, Guo G, Pei X, Wang X, Li L, Xiong Y, Qiu X (2019) Baicalin Inhibits cell viability, migration and invasion in breast cancer by regulating miR-338-3p and MORC4. Onco Targets Ther 12:11183–11193
Gattolliat CH, Uguen A, Pesson M, Trillet K, Simon B, Doucet L, Robaszkiewicz M, Corcos L (2015) MicroRNA and targeted mRNA expression profiling analysis in human colorectal adenomas and adenocarcinomas. Eur J Cancer 51:409–420
Hong Y, Chen X, Liang Z, Xu Z, Li Y, Pan Y (2020) MiR-338-3p inhibits cell migration and invasion in human hypopharyngeal cancer via downregulation of ADAM17. Anticancer Drugs 31:925–931
Huang S, Zhang X, Guan B, Sun P, Hong CT, Peng J, Tang S, Yang J (2019) A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am J Transl Res 11:2455–2462
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157
Jiang J, Lin H, Shi S, Hong Y, Bai X, Cao X (2020) Hsa_circRNA_0000518 facilitates breast cancer development via regulation of the miR-326/FGFR1 axis. Thorac Cancer 11:3181–3192
Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL (2017) Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 7:1566–1576
Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, Andrews R, Zhong W, Zhang X, Song E, Gong C (2019) Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 10:55
Ma F, Zhang L, Ma L, Zhang Y, Zhang J, Guo B (2017) MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J Exp Clin Cancer Res 36:158
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M (2013a) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N (2013b) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S (2017) Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform 18:780–788
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A (2016) Cancer treatment and survivorship statistics, 2016. A Cancer Journal for Clinicians 66:271–289
Pan G, Mao A, Liu J, Lu J, Ding J, Liu W (2020) Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif 53:12720
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777
Shen Y, Zhang M, Da L, Huang W, Zhang C (2020) Circular RNA circ_SETD2 represses breast cancer progression via modulating the miR-155-5p/SCUBE2 axis. Open Med (wars) 15:940–953
Sun F, Yu M, Yu J, Liu Z, Zhou X, Liu Y, Ge X, Gao H, Li M, Jiang X, Liu S, Chen X, Guan W (2018) miR-338-3p functions as a tumor suppressor in gastric cancer by targeting PTP1B. Cell Death Dis 9:522
Tang Q, Hann SS (2020) Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther 13:2067–2092
Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A (2010) FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Can Res 70:2085–2094
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM (2019) The landscape of circular RNA in cancer. Cell 176:869-881.e813
Wu Z, Wu J, Zhao Q, Fu S, Jin J (2020) Emerging roles of aerobic glycolysis in breast cancer. Clin Transl Oncol 22:631–646
Xie G, Ke Q, Ji YZ, Wang AQ, Jing M, Zou LL (2018) FGFR1 is an independent prognostic factor and can be regulated by miR-497 in gastric cancer progression. Braz J Med Biol Res 52:e7816
Yan L, Zheng M, Wang H (2019) Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Management and Research 11:1033–1041
Yedjou CG, Tchounwou PB, Payton M, Miele L, Fonseca DD, Lowe L, Alo RA (2017) Assessing the racial and ethnic disparities in breast cancer mortality in the United States. Int J Environ Res Public Health 14(5):486
Zhang L, Ding F (2019) Hsa_circ_0008945 promoted breast cancer progression by targeting miR-338-3p. Onco Targets Ther 12:6577–6589
Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL (2018) CircRNA: a novel type of biomarker for cancer. Breast Cancer 25:1–7
Zhang L, Yan R, Zhang SN, Zhang HZ, Ruan XJ, Cao Z, Gu XZ (2019) MicroRNA-338-3p inhibits the progression of bladder cancer through regulating ETS1 expression. Eur Rev Med Pharmacol Sci 23:1986–1995
Zhang J, Ke S, Zheng W, Zhu Z, Wu Y (2020) Hsa_circ_0003645 promotes breast cancer progression by regulating miR-139-3p/HMGB1 axis. Onco Targets Ther 13:10361–10372
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics Approval
Written informed consent was obtained from patients with approval by the Institutional Review Board in Xianning Central Hospital, The First Affiliated Hospital of Hubei University of Science and Technology.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2021_10102_MOESM1_ESM.tif
Supplementary file1 (TIF 66 kb) Knockdown of circ-TFF1 inhibited GLUT1 protein expression in BC cells. The protein expression of GLUT1 was detected by western blot assay in BT-549 and MDA-MB-231 cells transfected with si–N or si-circ-TFF1. ***P < 0.001.
Rights and permissions
About this article
Cite this article
Wan, L., Han, Q., Zhu, B. et al. Circ-TFF1 Facilitates Breast Cancer Development via Regulation of miR-338-3p/FGFR1 Axis. Biochem Genet 60, 315–335 (2022). https://doi.org/10.1007/s10528-021-10102-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10102-6