Abstract
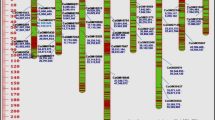
Understanding genetic diversity is very useful for scientific utilization for breeding. In this study, we estimated the genetic distances in a panel of 84 kenaf accessions collected from 26 countries and regions using ISSR markers. The results of UPGMA analysis showed that kenaf germplasm had abundant genetic variation, with genetic dissimilarity coefficients ranging from 0.01 to 0.62. The in-group dissimilarity coefficient (0.29) was observed in 84 kenaf accessions, and all the accessions could be divided into three groups: cultivars (L1–1), relatively wild species (L1–2 and L1–3), and wild species (the others). Further in-group analysis in group L1–1 (0.19) revealed that the kenaf cultivars could be divided into five subgroups with distinct regional characteristics. It is imperative that genes be exchanged among all kinds of tested varieties from different origins. The results provide a useful basis for kenaf germplasm research and breeding.


Similar content being viewed by others
References
Chen A, Li D, Tan H (2005) Analysis on kenaf cultivar genealogy and breeding tactics in China. Plant Fiber Prod 27(4):176–180
Chen M, Wei C, Qi J, Chen X, Su J, Li A, Tao A, Wu W (2011) Genetic linkage map construction for kenaf using SRAP, ISSR and RAPD markers. Plant Breed 130:79–687
Cheng Z, Lu B, Sameshima K, Chen J (2002) Comparative studies of genetic diversity in kenaf (Hibiscus cannabinus L.) varieties based on analysis of agronomic and RAPD data. Hereditas 136:231–239
Cheng Z, Yang X, Lu B, Jiao D, Chen J (2003) Genetic diversity and molecular identification of kenaf germplasm II, Diversity and genetic relationships of kenaf germplasm assessed by AFLP analysis. Plant Fiber Prod 25:162–167
Deng L, Su J, Huang P, Li A (1991) Morphological and taxonomic studies of kenaf germplasm. China’s Fiber Crops 4:16–20
Guo A, Zhou P, Su J (2002) Random amplified polymorphic DNA (RAPD) analyses among Hibiscus cannabinus and related species. J Trop Subtrop Bot 10:306–312
Li A, Deng L, Su J (1992) The preliminary identification of wild kenaf and relative species in East-Africa. J Anhui Agricul Sci 20:325–327
Li D, He X, Liu G, Huang B (2011) Genetic diversity and phylogenetic relationship of Tadehagi in southwest China evaluated by inter-simple sequence repeat (ISSR). Genet Resour Crop Evol 58:679–688
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Qi J, Li W, Wu W (1997) Studies on the origin and evolution of jute. Acta Agron Sin 23:677–682
Qi J, Li W, Wu W, Lin L, Lin P, Lu Q, Zheng Y (1999) Theory and practice of kenaf germplasm innovation. China’s Fiber Crops 21:33–44
Qi J, Fang P, Xu J, Lin L, Wu J (2007) High efficient utilization of jute/kenaf biomass and construction of industrial zone for territorial development. Plant Fiber Sci China 29:57–63
Rohlf F (2000) NTsys-pc: numerical taxonomy and multivariate analysis system. Version 2.1. Exeter Publishing, Setauket
Su J (2002) Strategies for protection and sustainable utilization of genetic diversity of bast fiber crops in China. J Plant Genet Resour 3:41–46
Su J, Deng L (1996) Study on the morphology and taxonomy of Hibiscus L. China’s Fiber Crops 18:18–20
Su J, Gong Y, Guan F, Guang F, Lu Y, Li Y, Dai Z (2003) The collection, conservation, regeneration and utilization of bast fiber crop germplasms. Plant Fiber Sci China 25:4–9
Xiong H (1989) Report of the germplasm collecting for jute and kenaf in Tanzania. China’s Fiber Crops 3:18–22
Xu J, Qi J, Fang P, Li A, Lin L, Wu J, Tao A (2007) Optimized CTAB protocol for extracting genomic DNA from kenaf and improved PCR amplifications of ISSR and SRAP. Plant Fiber Sci China 29:179–183
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
This project was sponsored by funds from the National Natural Science Foundation of China (30571188); the National Agri-Industry Technology Research System for Crops of Bast and Leaf Fiber, China (nycytx-19-E05); and the Construction of Germplasm Resources Platform for Bast Fiber Crops in Fujian (2010N2002).
Author information
Authors and Affiliations
Corresponding author
Additional information
Liwu Zhang and Aiqing Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, L., Li, A., Wang, X. et al. Genetic Diversity of Kenaf (Hibiscus cannabinus) Evaluated by Inter-Simple Sequence Repeat (ISSR). Biochem Genet 51, 800–810 (2013). https://doi.org/10.1007/s10528-013-9608-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-013-9608-7