Abstract
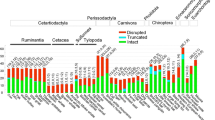
Bitter taste reception is expected to be associated with dietary selection and to prevent animals from ingesting potentially harmful compounds. To investigate the genetic basis of bitter taste reception, we reconfirmed the bitter taste receptor (T2R) genes from cow (herbivore) and dog (carnivore) genome sequences and identified the T2R repertoire from the draft genome of the bat (insectivore) for the first time using an automatic data-mining method. We detected 28 bitter receptor genes from the bat genome, including 9 intact genes, 8 partial but putative functional genes, and 9 pseudogenes. In the phylogenetic analysis, most of the T2R genes from the three species intermingle across the tree, suggesting that some are conserved among mammals with different dietary preferences. Furthermore, one clade of bat-specific genes was detected, possibly implying that the insectivorous mammal could recognize some species-specific bitter tastants. Evolutionary analysis shows strong positive selection was imposed on this bat-specific cluster, indicating that positive selection drives the functional divergence and specialization of the bat bitter taste receptors to adapt diets to the external environment.


Similar content being viewed by others
References
Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS (2000) A novel family of mammalian taste receptors. Cell 100:693–702
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Birney E, Clamp M, Durbin R (2004) GeneWise and Genomewise. Genome Res 14:988–995
Blum M (1981) Chemical defense of arthropods. Academic Press, New York
Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W (2002) The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet 32:397–401
Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ (2000) T2Rs function as bitter taste receptors. Cell 100:703–711
Eisner T (1970) Chemical defense against predators in arthropods. Academic Press, New York
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fenton M, Barclay R (1980) Myotis lucifugus. Mamm Species 142:1–8
Fenton M, Bell G (1979) Echolocation and feeding behaviour in four species of Myotis (Chiroptera). Can J Zool 57:1271–1277
Fischer A, Gilad Y, Man O, Paabo S (2005) Evolution of bitter taste receptors in humans and apes. Mol Biol Evol 22:432–436
Gilbertson TA, Damak S, Margolskee RF (2000) The molecular physiology of taste transduction. Curr Opin Neurobiol 10:519–527
Glendinning JI (1994) Is the bitter rejection response always adaptive? Physiol Behav 56:1217–1227
Glendinning JI, Smith JC (1994) Consistency of meal patterns in laboratory rats. Physiol Behav 56:7–16
Glendinning JI, Tarre M, Asaoka K (1999) Contribution of different bitter-sensitive taste cells to feeding inhibition in a caterpillar (Manduca sexta). Behav Neurosci 113:840–854
Go Y (2006) Proceedings of the SMBE Tri-National Young Investigators’ Workshop 2005. Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol Biol Evol 23:964–972
Go Y, Satta Y, Takenaka O, Takahata N (2005) Lineage-specific loss of function of bitter taste receptor genes in humans and nonhuman primates. Genetics 170:313–326
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217
Patterson B, Willig MR, Stevens RD (2003) Trophic strategies, niche partitioning, and patterns of ecological organization. University of Chicago press, Chicago
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shi P, Zhang J (2006) Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol 23:292–300
Shi P, Zhang J, Yang H, Zhang YP (2003) Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol Biol Evol 20:805–814
Simmons NB (2005) Mammal species of the world: a taxonomic and geographic reference. Johns Hopkins University Press, Baltimore, MD
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Weatherston J, Percy JE (1970) Arthropod defensive secretions. Academic Press, New York
Wong GT, Gannon KS, Margolskee RF (1996) Transduction of bitter and sweet taste by gustducin. Nature 381:796–800
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Wong WS, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Acknowledgments
This work was supported by grants under the Key Construction Program of the National “985” Project and “211” Project.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors Yingying Zhou and Dong Dong contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Y., Dong, D., Zhang, S. et al. Positive Selection Drives the Evolution of Bat Bitter Taste Receptor Genes. Biochem Genet 47, 207–215 (2009). https://doi.org/10.1007/s10528-008-9218-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-008-9218-y