Abstract
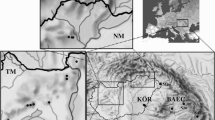
We analyzed a portion of mitochondrial COI gene sequences (658 bp) to investigate the genetic diversity and geographic variation of the swallowtail butterfly, Papilio xuthus L. (Lepidoptera: Papilionidae), and the cabbage butterfly, Pieris rapae L. (Lepidoptera: Pieridae). Papilio xuthus showed a moderate level of sequence divergence (0.91% at maximum) in 15 haplotypes, whereas Pi. rapae showed a moderate to high level of sequence divergence (1.67% at maximum) in 30 haplotypes, compared with other relevant studies. Analyses of population genetic structure showed that most populations are not genetically differentiated in both species. The distribution pattern of both species appears to be consistent with category IV of the phylogeographic pattern sensu Avise: a phylogenetic continuity, an absence of regional isolation of mtDNA clones, and extensive distribution of close clones. The observed pattern of genetic diversity and geographic variation of the two butterfly species seem to reflect the abundant habitats, abundant host plants, and flying abilities in connection with the lack of historical biogeographic barriers.



Similar content being viewed by others
References
Avise JC (1989) Gene trees and organismal histories: a phylogenetic approach to population biology. Evolution 43:1192–1208
Avise JC (1994) Molecular markers, natural history, and evolution. Chapman and Hall, New York
Avise JC, Helfman GS, Saunders NC, Hales LS (1986) Mitochondrial DNA differentiation in North Atlantic eels: population genetic consequences of an unusual life history pattern. Proc Natl Acad Sci USA 83:4350–4354
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987a) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18:489–522
Avise JC, Reeb CA, Sunders NC (1987b) Geographic population structure and species differences in mitochondrial DNA of mouthbrooding marine catfishes (Ariidae) and demersal spawning toadfishes (Batraxhoididae). Evolution 41:991–1002
Bae JS, Kim I, Kim SR, Jin BR, Shon HD (2001) Mitochondrial DNA sequence variation of the mushroom pest flies, Lycoelia mali (Diptera: Sciaridae), and, Coboldia fuscipes (Diptera: Scatopsidae), in Korea. Appl Entomol Zool Jpn 36:451–457
Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753
Ball RN Jr, Freeman S, James FC, Berminghan E, Avise JC (1988) Phylogeographic structue of red-winged blackbirds assessed by mitochondrial DNA. Proc Natl Acad Sci USA 85:1558–1562
Brower AVZ (1994) Phylogeny of Heliconius butterflies inferred from mitochondrial DNA sequences (Lepidoptera: Nymphalidae). Mol Phyl Evol 3:159–174
Ewens WJ (1990) Population genetic theory-the past and the future. In: Lessard S (ed) Mathematical and statistical development of evolutionary theory. Kluwer Academic Publishing, New York, pp 177–227
Excoffier L, Laval G, Schneider S (2005) Arlequin version 3.0. An integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Fauvelot C, Cleary FR, Menken BJ (2006) Short-term impact of disturbance on genetic diversity and structure of Indonesian populations of the butterfly Drupania theda in East Kalimantan. Mol Ecol 15:2069–2081
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primer for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Herbert PD (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci USA 103:968–971
Harper GL, Maclean N, Goulson D (2003) Microsatellite markers to assess the influence of population size, isolation and demographic change on the genetic structure of the UK butterfly Polyommatus bellargus. Mol Ecol 12:3349–3357
Herbert PD, Cywinska A, Ball SL, de Waard JR (2003) Biological identification through DNA barcodes. Proc Roy Soc Lond B 270:313–321
Holsinger KE, Mason-Gamer RJ (1996) Hierarchical analysis of nucleotide diversity in geographically structured populations. Genetics 142:629–639
Jones RE, Gilbert N, Guppy M, Nealis V (1980) Long-distance movement of Pieris rapae. J Am Ecol 49:629–642
Kim I, Phillips CJ, Monjeau JA, Birney EC, Noack K, Pumo DE, Sikes RS, Dole JA (1998) Habitat islands, genetic diversity, and gene flow in a Patagonian rodent. Mol Ecol 7:667–678
Kim I, Bae JS, Choi KH, Jin BR, Lee KR, Sohn HD (2000a) Haplotype diversity and gene flow of the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Yponomeutidae), in Korea. Korean J Appl Entomol 39:43–52
Kim I, Bae JS, Choi KH, Kim SR, Jin BR, Lee KR, Sohn HD (2000b) Mitochondrial DNA polymorphism, gene flow, and population genetic structure of diamondback moths, Plutella xylostella (Lepidoptera: Yponomeutidae), in southern Korea. Korean J Entomol 30:21–32
Kim I, Bae JS, Shon HD, Kang PD, Ryu KS, Shon BH, Jeong WB, Jin BR (2000c) Genetic homogeneity in the domestic silkworm, Bombyx mori, and phylogenetic relationship between B. mori and wild silkworm, B. mandarina, using mitochondrial COI gene sequences. Int J Indust Entomol 1:9–17
Kim I, Bae JS, Lee KS, Kim ES, Ryu KS, Yoon HJ, Jin BR, Moon BJ, Sohn HD (2003) Mitochondrial COI-gene sequence-based population genetic structure of the diamondback moth, Plutella xylostella, in Korea. Korean J Genet 25:155–179
Kim I, Cha SY, Kim MA, Lee YS, Lee KS, Choi YS, Hwang JS, Jin BR, Han YS (2007a) Polymorphism and genomic structure of the A + T-rich region of mitochondrial DNA in the oriental mole cricket, Gryllotalpa orientalis (Orthoptera: Gryllotalpidae). Biochem Genet 45:589–610
Kim JG, Kim I, Bae JS, Jin BR, Kim KY, Kim SE, Choi JY, Choi YC, Lee KY, Sohn HD, Noh SK (2001) Genetic subdivision of the firefly, Luciola lateralis (Coleoptera: Lampyridae), in Korea detected by mitochondrial COI gene sequences. Korean J Genet 23:25–33
Kim K-G, Jang SK, Park DW, Hong MY, Oh KH, Kim KY, Hwang JS, Han YS, Kim I (2007b) Mitochondrial DNA sequence variation of the tiny dragonfly, Nannophya pygmaea (Odonata: Libellulidae). Int J Indust Entomol 15:57–68
Kim YS (2002) Illustrated book of Korean butterflies in color. Kyo-Hak Pub. Co, Seoul, Korea
Lee SC, Kim I, Bae JS, Jin BR, Kim SE, Kim JK, Yang SR, Yoon HJ, Im SH, Sohn HD (2000) Mitochondrial DNA sequence variation of the firefly, Pyrocoelia rufa. Korean J Appl Entomol 39:181–191
Lee SC, Bae JS, Kim I, Yang WJ, Kim JK, Kim KY, Sohn HD, Jin BR (2003) Mitochondrial DNA sequence-based population genetic structure of the firefly, Pyrocoelia rufa (Coleoptera: Lampyridae). Biochem Genet 41:427–452
Li J, Choi YS, Kim I, Sohn HD, Jin BR (2004) Gnetic analysis of the diamondback moth, Plutella xylostella, collected from China using mitochondrial COI gene sequence. Int J Indust Entomol 9:137–144
Li J, Choi YS, Kim I, Sohn HD, Jin BR (2006) Genetic variation of the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Yponomeutidae) in China inferred from mitochondrial COI gene sequence. Eur J Entomol 103:605–611
Mun JH, Song YH, Heong KL, Roderick GK (1999) Genetic variation among Asian populations of rice planthoppers, Nilaparvata lugens and Sogatella furcifera (Hemiptera: Delphacidae): mitochondrial DNA sequences. Bull Entomol Res 89:245–253
Roderick GK (1996) Geographic structure of insect population: gene flow, phylogeography, and their uses. Annu Rev Entomol 41:263–290
Smith MA, Woodley NE, Janzen DH, Hallwachs W, Herbert PD (2006) DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci USA 103:3657–3662
Sperling FAH, Hickey DA (1994) Mitochondrial DNA sequence variation in the spruce budworm species complex (Choristoneura: Lepidoptera). Mol Biol Evol 11:656–665
Swofford DL (2002) PAUP: phylogenetic analysis using parsimony (and other methods). Version 4.10. Sinauer Associates, Sunderland, MA (on disk)
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:173–216
Acknowledgment
This work was supported by a grant (Code 2007040134004) from the Biogreen 21 Program, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jeong, H.C., Kim, J.A., Im, H.H. et al. Mitochondrial DNA Sequence Variation of the Swallowtail Butterfly, Papilio xuthus, and the Cabbage Butterfly, Pieris rapae . Biochem Genet 47, 165–178 (2009). https://doi.org/10.1007/s10528-008-9214-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-008-9214-2