Abstract
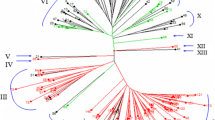
Genetic diversity and population genetic structure of autotetraploid and diploid populations of rice collected from Chengdu Institute of Biology, Chinese Academy of Sciences, were studied based on 36 microsatellite loci. Among 50 varieties, a moderate to high level of genetic diversity was observed at the population level, with the number of alleles per locus (A e) ranging from 2 to 6 (mean 3.028) and polymorphism information content ranging from 0.04 to 0.76 (mean 0.366). The expected heterozygosity (H e) varied from 0.04 to 0.76 (mean 0.370) and Shannon’s index (I) from 0.098 to 1.613 (mean 0.649). The autotetraploid populations showed slightly higher levels of A e, H e, and I than the diploid populations. Rare alleles were observed at most of the simple sequence repeat loci in one or more of the 50 accessions, and a core fingerprint database of the autotetraploid and diploid rice was constructed. The F-statistics showed genetic variability mainly among autotetraploid populations rather than diploid populations (F st = 0.066). Cluster analysis of the 50 accessions showed four major groups. Group I contained all of the autotetraploid and diploid indica maintainer lines and an autotetraploid and its original diploid indica male sterile lines. Group II contained only the original IR accessions. Group III was more diverse than either Group II or Group IV, comprising both autotetraploid and diploid indica restoring lines. Group IV included a japonica cluster of the autotetraploid and diploid rices. Furthermore, genetic differences at the single-locus and two-locus levels, as well as components due to allelic and gametic differentiation, were revealed between autotetraploid and diploid varieties. This analysis indicated that the gene pools of diploid and autotetraploid rice were somewhat dissimilar, as variation exists that distinguishes autotetraploid from diploid rices. Using this variation, we can breed new autotetraploid varieties with some important agricultural characters that were not found in the original diploid rice varieties.


Similar content being viewed by others
References
Anderson JA, Churchill GA, Sutrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linksion was different from both the indica and japonica age maps. Genome 36:181–186
Bingham ET, Groose RW, Woodfield DR, Kidwell KK (1994) Complementary gene interactions in alfalfa is greater in autotetraploids than in diploids. Crop Sci 34:823–829
Bussell JD (1999) The distribution of random amplified polymorphic DNA (RAPD) diversity among populations of Isotoma petraea (Lobeliaceae). Mol Ecol 8:775–789
Chen XM, Gao SL, Bian YY (2003) Induction and identification of autotetraploid of Mractylodes macrocephala and comparison of its peroxidase and diploids. J Plant Resour Environ 12(1):16–20 (Chinese with English abstract)
Guo M, Davis D, Birchler JA (1996) Dosage effects on gene expression in a maize ploidy series. Genetics 1424:l349–1355
Guo XP, Qin RZ, Chen X (2002) Factors affecting production of autotetraploid rice. J Plant Genet Resour 3(3):30–33
Hanson RE, Islam-Faridi MN, Crane CF, Zwick MS, Czeschin DG, Wendel JF, Mcknight TD, Price HJ, Stelly DM (1999) Ty1-copia-retrotransoson behavior in a polyploid cotton. Chromosome Res 8:73–76
Hovmalm HAP, Jeppsson N, Bartish IV, Nybom H (2004) RAPD analysis of diploid and tetraploid population of Aronia points to different reproductive strategies within the genus. Hereditas 141:301–312
Lee HS, Chen ZJ (2001) Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA 98:6753–6758
Liu B, Wendel JF (2003) Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol 29(3):365–379
Liu HJ, Wang H, Gao LH (1997) Comparisons of isoen-zymes between autotetraploid and diploid Chinese cabbage varieties of different ecotypes. J Nanjing Agric Univ 20(3):26–30 (Chinese with English abstract)
Luo ZW, Hackett CA, Bradshaw JE, McNicol JW, Milbourne D (2001) Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics (Bethesda) 157:1369–1382
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and application in rice genetics and breeding. Plant Mol Breed 35:89–99
Nayar NM (1981) Original and cytogenetics of rice. Agronomy Press, Beijing
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M, Li WH (1979) Mathematical mode for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76(10):5269–5273
Ni J, Colowit PM, Mackill D (2002) Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci 42:601–607
Pao WK, Qin RZ, Wu DY, Chen ZY, Song WC, Zhang YH (1985) High yielding tetraploid rice clones. Sci Agric Sin 6:64–66 (Chinese with English abstract)
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey SV, Rafalski A (1996) Comparison of RFLP, RAPD, AFLP and SSR (microsatellites) markers for germplasm analysis. Mol Breed 2:225–238
Rohlf FJ (1994) NTSys-pc, numerical taxonomy and multivariate analysis system, ver. 1.80. Exeter Software, New York
Roy JK, Lakshmikumaran MS, Balyan HS, Gupta PK (2004) AFLP-based genetic diversity and its comparison with diversity based on SSR, SAMPL, and phenotypic traits in bread wheat. Biochem Genet 42:43–59
Saghai MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Song ZP, Xu X, Wang B, Chen JK, Lu BR (2003) Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theor Appl Genet 107:1492–1499
Suh HS, Sato YI, Morishima H (1997) Genetic characterization of weedy rice (Oryza sativa L.) based on morphophysiology, isozymes and RAPD markers. Theor Appl Genet 94:316–321
Sunita J, Rajinder KJ, Susan R, McCouch SR (2004) Genetic analysis of Indian aromatic and quality rice (Oryza sativaL.) germplasm using panels of fluorescently-labeled microsatellite markers. Theor Appl Genet 109:965–977
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch SR (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1141–1451
Temnykh S, Park WD, Ayres NM, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Tu SB, Kong FL, Xu QF, He T (2003) Breakthrough in hybrid rice breeding with autotertraploid. CAS Bull 6:426–428 (in Chinese with English abstract)
Tu SB, Luan L, Liu YH, Long WB, Kong FL, He T, Xu QF, Yan WG, Yu MQ (2007) Production and heterosis analysis of rice (Oryza sativa L.) autotetraploid hybrids. Crop Sci (in press)
Wang ZW, Yu MD, Lu C (2002) The AFLP analysis of genetic diversity between diploid and artificially induced ho-mologous tetraploid of mulberry. Chin Bull Bot 19:194–200
Weir BS (1990) Genetic data analysis. Sinauer Associations, Sunderland, MA
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42(1):225–249
Xiao MY, Qing Z, Zeng QQ, Shan L, Gui FZ (2006) Analysis of genetic diversity and population differentiation of Larix potaninii var. chinensis using microsatellite DNA. Biochem Genet 44:491–501
Xie XQ, Gao SL 2002) The analysis of peroxidase iso-exnzyme electrophoresis and activity of ascorbate peroxi-dase(ASP) in Scutellaria baicalensis Georgi. J Plant Resour Environ 11:5–8 (Chinese with English abstract)
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) PopGene, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Canada
Yu SB, Xu WJ, Vijayakumar CHM, Ali J, Fu BY, Xu JL, Jiang YZ, Marghirang R, Domingo J, Aquino C, Virmani SS, Li ZK (2003) Molecular diversity and multilocus organization of the parental lines used in the international rice molecular breeding program. Theor Appl Genet 108:131–140
Zhao G, Tang Y (1994) Comparative study on main characters of a new strain of autotetraploid and its autodiploid parent stock in Tartar buckwheat. Bull Sci 5:321–325
Acknowledgments
The authors are grateful to Mr. Ye Yan, Chaolong Gao, and Chuanmin Zeng for their excellent technical assistance. The authors are also grateful to the editor and anonymous reviewers and professional colleagues for critical comments and help on the presentation of the manuscript. Also, special thanks go to the Chinese Academy of Sciences for funding this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luan, L., Wang, X., Long, WB. et al. Microsatellite Analysis of Genetic Variation and Population Genetic Differentiation in Autotetraploid and Diploid Rice. Biochem Genet 46, 248–266 (2008). https://doi.org/10.1007/s10528-008-9156-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-008-9156-8