Abstract
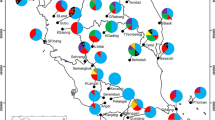
The East Himalaya-Hengduan Mountains region is the center of diversity of the genus Primula, and P. sikkimensis is one of the most common members of the genus in the region. In this study, the genetic diversity and structure of P. sikkimensis populations in China were assessed using inter-simple sequence repeat (ISSR) and chloroplast microsatellite markers. The 254 individuals analyzed represented 13 populations. High levels of genetic diversity were revealed by ISSR markers. At the species level, the expected heterozygosity and Shannon’s index were 0.4032 and 0.5576, respectively. AMOVA analysis showed that 50.3% of the total genetic diversity was partitioned among populations. Three pairs of chloroplast microsatellite primers tested yielded a total of 12 size variants and 15 chloroplast haplotypes. Strong cpDNA genetic differentiation (G ST = 0.697) and evidence for phylogeographic structure were detected (N ST = 0.788, significantly higher than G ST). Estimated rates of pollen-mediated gene flow are approximately 27% greater than estimated rates of seed-mediated gene flow in P. sikkimensis. Both seed and pollen dispersal, however, are limited, and gene flow among populations appears to be hindered by the patchiness of the species’ habitats and their geographic isolation. These features may have played important roles in shaping the genetic structure of P. sikkimensis. A minimum-spanning tree of chloroplast DNA haplotypes was constructed, and possible glacial refugia of P. sikkimensis were identified.



Similar content being viewed by others
References
Antrobus S, Lack AJ (1993) Genetics of colonizing and established populations of Primula veris. Heredity 71:252–258
Birky CW Jr, Maruyama T, Fuerst P (1983) An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics 103:513–527
Cahalan CM, Gliddon C (1985) Genetic neighbourhood sizes in Primula vulgaris. Heredity 54:65–70
Chung SM, Decker-Walters DS, Staub JE (2003) Genetic relationships within the Cucurbitaceae as assessed by consensus chloroplast simple sequence repeats (ccSSR) marker and sequence analyses. Can J Bot 81:814–832
Doyle J (1991) DNA protocols for plants-CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds) Molecular techniques in taxonomy. Springer, Berlin, pp 283–293
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plants populations. Heredity 72:250–259
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (1993) Phylip, Phylogenetic inference package, version 3.5.7. A computer program distributed by the author, http://www.evolution.genetics.washington.edu/phylip.html. Department of genetics, University of Washington, Seattle
Fontaine C, Lovett PN, Sanou H, Maley J (2004) Genetic diversity of the shea tree (Vitellaria paradoxa C.F. Gaertn), detected by RAPD and chloroplast microsatellite markers. Heredity 93:639–648
Ge XJ, Zhang LB, Yuan YM, Hao G, Chiang TY (2005) Strong genetic differentiation of the East-Himalayan Megacodon stylophorus (Gentianaceae) detected by Inter-Simple Sequence Repeats (ISSR). Biodivers Conserv 14:849–861
Glover BJ, Abbott RJ (1995) Low genetic diversity in the Scotish endemic Primula scotica Hook. New Phytol 129:147–153
Gupta M, Chyi YS, Romero-Severson J, Owen JL (1994) Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theor Appl Genet 89:998–1006
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer Associates, Sunderland, pp 43–63
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B 351:1291–1298
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New For 6:95–124
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hu CM (1994) On the geographical distribution of the Primulaceae. J Trop Subtrop Bot 2:1–14
Hu CM, Kelso S (1996) Primulaceae. In: Wu CY, Raven PH (eds) Flora of China, vol 15. Science Press, Beijing, China, and Missouri Botanical Garden, St. Louis, USA, pp 39–189
Ishihama F, Ueno S, Tsumura Y, Washitani I (2006) Effects of density and floral morph on pollen flow and seed reproduction of an endangered heterostylous herb, Primula sieboldii. J Ecol 94:846–855
Jacquemyn H, Honnay O, Galbusera P, Roldán-Ruiz I (2004) Genetic structure of the forest herb Primula elatior in a changing landscape. Mol Ecol 13:211–219
Kumar S, Tamura K, Nei M (2004) MEGA 3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398
Li XW, Li J (1993) A preliminary floristic study on the seed plants from the region of Hengduan Mountain. Acta Bot Yunnanica 15:217–231
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Ann Rev Ecol Syst 15:65–95
Lu Z, Wang YH, Peng YH (2006) Genetic diversity of Populus cathayana populations in southwestern China revealed by ISSR markers. Plant Sci 170:407–412
Miller MP (1997) Tools for population genetic analysis, ver. 1.3. Department of Biological Sciences, Northern Arizona University, Flagstaff, Arizona
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nan P, Peng SL, Shi SH, Ren H, Yang J, Zhong Y (2003a) Interpopulation congruence in Chinese Primula ovalifolia revealed by chemical and molecular markers using essential oils and ISSRs. Z Naturforsch 58:57–61
Nan P, Shi SH, Peng SL, Tian CJ, Zhong Y (2003b) Genetic diversity of Primula obconica from central and southwest China as revealed by ISSR markers. Ann Bot 91:329–333
Newton AC, Allnutt TR, Dvorak WS, Del Castillo RF, Ennos RA (2002) Patterns of genetic variation in Pinus chiapensis, a threatened Mexican pine, detected by RAPD and mitochondrial DNA RFLP markers. Heredity 89:191–198
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Peng YH, Lu ZX, Chen K, Luukkanen O, Korpelainen H, Li CY (2005) Population genetic survey of Populus cathayana originating from Southeastern Qinghai-Tibetan plateau of China based on SSR markers. Silvae Genet 54:116–122
Petit RJ (1999) Diversité Génétique et Histoire des Populations d’Arbres Forestiers. Dossier D’habilitation à Diriger Des Recherches. Université de Paris-Sud, Université Formation de Recherche Scientifique d’Orsay, Paris
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA (1995) Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci USA 92:7759–7763
Rao RR (1994) Biodiversity in India: Floristic Aspects. Bisen Singh Mahendra Pal Singh, Dehra Dun
Raspé O, Saumitou-Laprade P, Cuguen J, Jacquemart A-L (2000) Chloroplast DNA haplotype variation and population differentiation in Sorbus aucuparia L. (Rosaceae: Maloideae). Mol Ecol 9:1113–1122
Reisch C, Anke A, Röhl M (2005) Molecular variation within and between ten populations of Primula farinosa (Primulaceae) along an altitudinal gradient in the northern Alps. Basic Appl Ecol 6:35–45
Richards J (2002). Primula, New edition. BT Batford, London
Schneider S, Roessli D, Excoffier L (2000) Arlequin, Version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Sun H (2002) Evolution of Arctic-tertiary flora in Himalayan–Hengduan Mountains. Acta Bot Yunnanica 24:671–688
Syamsuardi, Okada H (2002) Genetic diversity and genetic structure of populations of Ranunculus japonicus Thunb. (Ranunculaceae). Plant Species Biol 17:59–69
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Takhtajan A (1969) Flowering plants, origin and dispersal. Oliver and Boyd, Edinburgh
Van Rossum F, De Sousa SC, Triest L (2004) Genetic consequences of habitat fragmentation in an agricultural landscape on the common Primula veris, and comparison with its rare congener, P. vulgaris. Conserv Genet 5:231–245
Van Rossum F, Triest L (2003) Spatial genetic structure and reproductive success in fragmented and continuous populations of Primula vulgaris. Folia Geobot 38:239–254
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002). Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Viard F, El-Kassably YA, Ritland K (2001) Diversity and genetic structure in populations of Pseudotsuga menziesii (Pinaceae) at chloroplast microsatellite loci. Genome 44:336–344
Weising K, Gardner RC (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Wolfe AD, Liston A (1998) Contributions of PCR-based methods to plant systematics and evolutionary biology. In: Soltis DE, Soltis PS, Doyle JJ (eds) Plant molecular systematics II. Kluwer, Boston, pp 43–86
Wu ZY (1988) Hengduan mountain flora and her significance. J Jpn Bot 63:297–311
Wu ZY, Wu SG (1996) A proposal for new floristic kingdom (realm). In: Zhang AL, Wu SG (eds) Floristic characteristics and diversity of east Asian plants. Springer-Verlag, Hongkong, China Higher Education Press, Beijing, pp 3–42
Xia T, Chen SL, Chen SY, Ge XJ (2005) Genetic variation within and among populations of Rhodiola alsia (Crassulaceae) native to the Tibetan plateau as detected by ISSR makers. Biochem Genet 43:87–101
Xue DW, Ge XJ, Hao G, Zhang CQ (2004) High genetic diversity in a rare, narrowly endemic primrose species: Primula interjacens (Primulaceae) by ISSR analysis. Acta Bot Sin 46:1163–1169
Yeh FC, Yang R, Boyle T (1999) PopGene. Microsoft Windows-based freeware for population genetic analysis. Release 1.31. University of Alberta, Edmonton
Zhang RZ, Zheng D, Yang QY, Liu YH (1997) Physical geography of Hengduan mountains. Science Press, Beijing
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
This project was financially supported by the National Natural Science Foundation of China (Nos. 30470125, 40671066) and the Natural Science Foundation of Guangdong Province (No. 31255).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, FY., Ge, XJ., Gong, X. et al. Strong Genetic Differentiation of Primula sikkimensis in the East Himalaya–Hengduan Mountains. Biochem Genet 46, 75–87 (2008). https://doi.org/10.1007/s10528-007-9131-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-007-9131-9