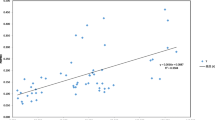
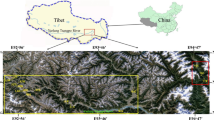
ISSR markers were used to analyze the genetic diversity and genetic structure of eight natural populations of Cupressus chengiana in China. ISSR analysis using 10 primers was carried out on 92 different samples. At the species level, 136 polymorphic loci were detected. The percentage of polymorphic bands (PPB) was 99%. Genetic diversity (H e) was 0.3120, effective number of alleles (A e) was 1.5236, and Shannon’s information index (I) was 0.4740. At the population level, PPB = 48%, A e=1.2774, H e=0.1631, and I=0.2452. Genetic differentiation (G st) detected by Nei’s genetic diversity analysis suggested 48% occurred among populations. The partitioning of molecular variance by AMOVA analysis indicated significant genetic differentiation within populations (54%) and among populations (46%; P < 0.0003). The average number of individuals exchanged between populations per generation (N m ) was 0.5436. Samples from the same population clustered in the same population-specific cluster, and two groups of Sichuan and Gansu populations were distinguishable. A significantly positive correlation between genetic and geographic distance was detected (r=0.6701). Human impacts were considered one of the main factors to cause the rarity of C. chengiana, and conservation strategies are suggested based on the genetic characters and field investigation, e.g., protection of wild populations, reestablishment of germplasm bank, and reintroduction of more genetic diversity.


Similar content being viewed by others
REFERENCES
Bai, D., Brandle, J., and Reeleder, R. (1997). Genetic diversity in North American ginseng (Panax quinquefolius L.) grown in Ontario detected by RAPD analysis. Genome 40:111–115.
Bartel, J. A., Adams, R. P., James, S. A., Mumba, L. E., and Pandey, R. N. (2003). Variation among Cupressus species from the western hemisphere based on random amplified polymorphic DNAs. Biochem. Systemat. Ecol. 31:693–702.
Buchert, G. P., Rajora, O. P., and Hood, J. V. (1997). Effects of harvesting on genetic diversity in old growth Eastern white pine in Ontario, Canada. Conserv. Biol. 11(3):747–758.
Chen, X. Y. (2000). Effects of habitat fragmentation on genetic structure of plant populations and implications for the biodiversity conservation. Acta Ecol. Sin. 20(3):884–892.
El-Kassaby, Y. A. (1991). Genetic variation within and among conifer populations: Review and evaluation of methods. In Fineschi, S., Malvolti, M. E., Cannata, F., and Hattemer, H. H. (eds.), Biochemical Markers in the Population Genetics of Forest Trees, Academic Publishing, S. P. B., The Hague, pp. 61–76.
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Applications to human mitochondrial DNA restriction data. Genetics 131:479–491.
Fernández, M. E., Figueiras, A. M., and Benito, C. (2002). The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification, and genetic diversity among barley cultivars with known origin. Theor. Appl. Genet. 104:845–851.
Frankham, R., Ballou, J. D., and Briscoe, D. A. (2002). Introduction to Conservation Genetics, Cambridge University Press, Cambridge, UK.
Fu, L.-G. (1992). Red Book of China: Rare and Threatened Plants, Science Publishing House Press, Beijing.
Fu, Y., and Dane, F. (2003). Allozyme variation in endangered Castanea pumila var. pumila. Ann. Bot. 92:223–230.
Ge, S., Zhang, D. M., Wang, H. Q., and Rao, G. Y. (1997). Allozyme variation in Ophiopogon xylorrhizus, an extreme endemic species of Yunnan, China. Conserv. Biol. 11(2):562–565.
Ge, S. (1988). Isozymes and studies on population genetic variation in forest trees. J. Nanjing Forest. Univ. 1:68–77.
Ge, S., Hong, D.-Y., Wang, H.-Q., Liu, Z.-Y., and Zhang, C.-M. (1998). Population genetic structure and conservation of an endangered conifer, Cathaya argyrophylla (Pinaceae). Int. J. Plant Sci. 159(2):351–357.
Ge, Y. Q., Qiu, Y. X., Ding, B. Y., and Fu, C. X. (2003). An ISSR analysis on population genetic diversity of the relict plant Ginkgo Biloba. Biodivers. Sci. 11(4):276–287.
Godt, M. J. W., Hamrick, J. L., and Meier, A. (2004). Genetic diversity in Cymophyllus fraserianus (Cyperaceae), a rare monotypic genus. Genetica 122(2):207–215.
Hamrick, J. L., and Godt, M. J. W. (1990). Allozyme diversity in plant species. In Brown, A. H. D., Clegg, M. T., Kahler, A. L., and Wier, B. S. (eds.), Plant Population Genetics, Breeding, and Genetic Resources, Sinauer Associates, Sunderland, MA, pp. 43–63.
Hamrick, J. L., Linhart, Y. B., and Mitton, J. B. (1979). Relationships between life history characteristics and electrophoretically detectable genetic variance in plants. Annu. Rev. Ecol. System. 10:173–200.
Hodkinson, T. R., Chase, M. W., and Renvoize, S. A. (2002). Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropgoneae, Poaceae) using AFLP and ISSR PCR. Ann. Bot. 89:627–636.
Hogbin, P. M., Peakall, R., and Sydes, M. A. (2000). Achieving practical outcomes from genetic studies of rare Australian plants. Aust. J. Bot. 48:375–382.
Holsinger, K. E., and Gottlieb, L. D. (1991). Conservation of rare and endangered plants: Principles and prospects. In Falk, D. A., and Holsinger, K. E. (eds.), Genetics and Conservation of Rare Plants, Oxford University Press, Oxford, pp. 195–223.
Kjolner, S., Sastad, S. M., Taberlet, P., and Brochmann, C. (2004). Amplified fragment length polymorphism versus random amplified polymorphic DNA markers: Clonal diversity in Saxifraga cernua. Mol. Ecol. 13:81–86.
Laurence, G., Hu, Z.-L., and Zavarin, E. (1998). Foliage terpenoids of Chinese Cupressus species. Biochem. System. Ecol. 26:899–913.
Li, L.-C., and Fu, Y.-X. (1996). Studies on the karyotypes and the cytogeography of Cupressus (Cupressaceae). Acta Phytotaxonom. Sin. 34(2):117–123.
Li, X. D., Huang, H. W., and Li, J. Q. (2003). Genetic diversity of the relict plant Metasequoia glyptostroboides. Biodivers. Sci. 11(2):100–108.
Liu, X. L., Mo, C. L., Xiang, C. H., and Shu, Y. M. (2001). Natural characteristics, reforestation, and reconstruction in dry valley in western Sichuan. J. Sichuan Forest. Sci. Technol. 22(2):10–17.
Loveless, M. P., and Hamrick, J. L. (1984). Ecological determinant of genetic structure in plant populations. Ann. Rev. Ecol. System. 15:65–95.
Maki, M., and Horie, S. (1999). Random amplified polymorphic DNA (RAPD) markers revealed less genetic variation in the endangered plant Cerastium fisherianum var. molle than in the widespread conspecific C. fischerianum var. fischerianum (Caryophyllaceae). Mol. Ecol. 8:145–150.
Mattioni, C., Casasoli, M., Gonzalez, M., Ipinza, R., and Villani, F. (2002). Comparison of ISSR and RAPD markers to characterize three Chilean Nothofagus species. Theor. Appl. Genet. 104:1064–1070.
Neel, M. C., and Ellstrand, N. C. (2003). Conservation of genetic diversity in the endangered plant Eriogonum ovalifolium var. vineum (Polygonaceae). Conserv. Genet. 4(3):337–352.
Nei, M. (1972). Genetic distance between populations. Am. Nat. 106:283–292.
Rohlf, F. J. (1998). NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, ver. 2.02, Exeter Software, Setauket, NY.
Rushforth, K., Adams, R. P., Zhong, M., Ma, X., and Pandey, R. N. (2003). Variation among Cupressus species from the eastern hemisphere based on Random Amplified Polymorphic DNAs (RAPDs). Biochem. System. Ecol. 31:17–24.
Sabrina, S., and Sabri, S. (1999). Genetic diversity in natural Cupressus sempervirens L. populations in Turkey. Biochem. System. Ecol. 27:799–814.
Schonswetter, P., Tribsch, A., and Niklfeld, H. (2004). Amplified Fragment Length Polymorphism (AFLP) reveals no genetic divergence of the Eastern Alpine endemic Oxytropis campestris subsp. tiroliensis (Fabaceae) from widespread subsp. campestris. Plant Syst. Evol. 244:245–255.
Shi, D., and Wang, M. (1994). The studies on the karyotype and the relationship of the two endemic species of genus Cupressus in China. J. Sichuan Agric. Univ. 12(1):92–96.
Shukla, N., Sangwan, N. S., Misra, H. O., and Sangwan, R. S. (2003). Genetic diversity analysis in Boerhavia diffusa L. of different geographic locations in India using RAPD markers. Genet. Resourc. Crop Evol. 50(6):587–601.
Slatkin, M. (1985). Gene flow in natural populations. Annu. Rev. Ecol. System. 16:393–430.
Slatkin, M. (1987). Gene flow and the geographic structure of natural populations. Science 236:787–792.
Sun, M., Wong, K. C., and Lee, J. S. Y. (1998). Reproductive biology and population genetic structure of Kandelia candel (Rhizophoraceae), a viviparious mangrove species. Am. J. Bot. 85:1631–1637.
Wang, T., Su, Y. J., Li, X. Y., Zheng, B., Chen, G. P., and Wang, B. S. (2003). RAPD analysis of the genetic variation within populations of a relict tree fern, Alsophila spinulosa (Cyatheaceae). Acta Ecol. Sin. 23(6):1200–1205.
Wang, X. Q., Zou, Y. P., Zhang, D. M., and Hong, D. Y. (1996). The RAPD for the genetic diversity of Cathaya argyrophylla. Sci. China 26(5):436–441.
Wang, Y., Tang, S. Q., and Li, X. K. (2004). The genetic diversity of the endangered plant Abies yuanbaoshanensis. Biodivers. Sci. 12(2):269–273.
Weising, K., Nybom, H., Wolff, K., and Meyer, W. (1995). DNA Fingerprints in Plants and Fungi, CRC Press, Boca Raton, FL.
Wilson, E. H. (1916). An enumeration of the woody plants collected in western China for the Arnold Arboretum of Harvard University during the years 1907, 1908, and 1910, by E. H. Wilson. Cambridge University Press, Cambridge, UK, pp. 54–55.
Wright, S. (1931). Evolution in Mendelian population. Genetic 16:97–159.
Wright, S. (1951). The genetical structure of populations. Ann. Eugen 15:323–354.
Yeh, F. C., Yang, R. C., and Boyle, T. (1997). Popgene, the User Friendly Shareware for Population Genetic Analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton, Canada.
Yuan, Z. Z., Bao, W. K., and He, B. H. (2004a). Life table and survival analysis of Cupressus chengiana population in western Sichuan, China. Acta Botan. Yunnanica 26(4):373–381.
Yuan, Z. Z., Bao, W. K., and He, B. H. (2004b). Dynamic analysis of age structure of Cupressus chengiana populations in four geographical areas. J. Plant Res. Environ. 13(3):25–30.
Zhang, M., and Wang, Q. M. (1987). Primary investigation of the germination properties of 15 kinds of cypress seeds. Jiangshu Forest. Technol. 2:8–9.
Zheng, W. J., and Fu, G. L. (1978). Flora of China, Vol. 7, Science Press, Beijing,pp. 328–337.
Zietkiewicz, E., Rafalski, A., and Labuda, D. (1994). Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction. Genomics 20:176–183.
Zou, Y.-P., Ge, S., and Wang, X.-D. (2001). Molecular Markers used in Systematics and Development Botany, Science Press, Beijing.
ACKNOWLEDGMENTS
This research was funded by the “Studies on endangered mechanism and conservation of Cupressus chengiana” of the Important Direction Project of China Academic of Science, Project contract number KSCX2-SW-104. We are very grateful to our colleagues at Chengdu Institute of Botany, CAS, for assistance with sample collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hao, B., Li, W., Linchun, M. et al. A Study of Conservation Genetics in Cupressus chengiana, an Endangered Endemic of China, Using ISSR Markers. Biochem Genet 44, 29–43 (2006). https://doi.org/10.1007/s10528-006-9011-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-006-9011-8