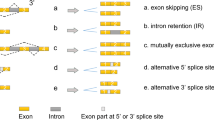
Regulation of alternative splicing is carried out by RNA-binding proteins. Each alternative splicing event is controlled by several RNA-binding proteins, which in combination create the distribution of alternative splicing products in a given cell type. Transmembrane protein CD44 plays an important role at various stages of the metastatic cascade and is considered as a promising molecule for the therapy of tumor diseases and the construction of prognostic classifiers. However, the functions of specific isoforms of this protein may differ significantly. In this work, we performed a bioinformatic search of RNA-binding proteins that can determine the expression of clinically significant isoforms 3 and 4 of CD44 protein. The analysis revealed five RNA-binding proteins, three of which (OAS1, ZFP36L2, and DHX58) are shown for the first time as potential regulators of the studied process.
Similar content being viewed by others
References
Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4(1):33-45. https://doi.org/10.1038/nrm1004
Wang Z, Zhao K, Hackert T, Zöller M. CD44/CD44v6 a reliable companion in cancer-initiating cell maintenance and tumor progression. Front. Cell Dev. Biol. 2018;6:97. https://doi.org/10.3389/fcell.2018.00097
Chen J, Zhou J, Lu J, Xiong H, Shi X, Gong L. Significance of CD44 expression in head and neck cancer: a systemic review and meta-analysis. BMC Cancer. 2014;14:15. https://doi.org/10.1186/1471-2407-14-15.
Wang Z, Tang Y, Xie L, Huang A, Xue C, Gu Z, Wang K, Zong S. The prognostic and clinical value of CD44 in colorectal cancer: a meta-analysis. Front. Oncol. 2019;9:309. https://doi.org/10.3389/fonc.2019.00309
Hong I, Hong SW, Chang YG, Lee WY, Lee B, Kang YK, Kim YS, Paik IW, Lee H. Expression of the cancer stem cell markers CD44 and CD133 in colorectal cancer: an immunohistochemical staining analysis. Ann. Coloproctol. 2015;31(3):84-91. https://doi.org/10.3393/ac.2015.31.3.84
Azevedo R, Gaiteiro C, Peixoto A, Relvas-Santos M, Lima L, Santos LL, Ferreira JA. CD44 glycoprotein in cancer: a molecular conundrum hampering clinical applications. Clin. Proteomics. 2018;15:22. https://doi.org/10.1186/s12014-018-9198-9
Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl Acad. Sci. USA. 1992;89(24):12160-12164. https://doi.org/10.1073/pnas.89.24.12160
Ruiz P, Schwärzler C, Günthert U. CD44 isoforms during differentiation and development. Bioessays. 1995;17(1):17-24. https://doi.org/10.1002/bies.950170106
Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 2011;121(3):1064-1074. https://doi.org/10.1172/JCI44540
Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, Deng Y, Hu X, Zhang J, Gao XD, Kang Y, Mercurio AM, Goel HL, Cheng C. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019;33(3-4):166-179. https://doi.org/10.1101/gad.319889.118
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-140. https://doi.org/10.1093/bioinformatics/btp616
Giudice G, Sánchez-Cabo F, Torroja C, Lara-Pezzi E. ATtRACT-a database of RNA-binding proteins and associated motifs. Database (Oxford). 2016;2016:baw035. https://doi.org/10.1093/database/baw035
Giulietti M, Piva F, D’Antonio M, D’Onorio De Meo P, Paoletti D, Castrignanò T, D’Erchia AM, Picardi E, Zambelli F, Principato G, Pavesi G, Pesole G. SpliceAid-F: a database of human splicing factors and their RNA-binding sites. Nucleic Acids Res. 2013;41(Database issue):D125-D131. https://doi.org/10.1093/nar/gks997
Novosad VO, Polikanova IS, Tonevitsky EA, Maltseva DV. Expression of CD44 isoforms in tumor samples and cell lines of human colorectal cancer. Bull. Exp. Biol. Med. 2022;173(1):155-159. https://doi.org/10.1007/s10517-022-05512-4
Waskom ML. Seaborn: statistical data visualization. J. Open Source Softw. 2021. https://doi.org/10.21105/joss.03021
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, Miyazawa K. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J. Biol. Chem. 2014;289(40):27386-27399. https://doi.org/10.1074/jbc.M114.589432
Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, Rousset F, Tazi J, Abou Elela S, Chabot B. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell. Biol. 2013;33(2):396-405. https://doi.org/10.1128/MCB.01174-12
Di C, Syafrizayanti, Zhang Q, Chen Y, Wang Y, Zhang X, Liu Y, Sun C, Zhang H, Hoheisel JD. Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ. 2019;26(7):1181-1194. https://doi.org/10.1038/s41418-018-0231-3
Samatov TR, Tonevitsky AG, Schumacher U. Epithelial-mesenchymal transition: focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Mol. Cancer. 2013;12(1):107. https://doi.org/10.1186/1476-4598-12-107
Senbanjo LT, Chellaiah MA. CD44: a multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell Dev. Biol. 2017;5:18. https://doi.org/10.3389/fcell.2017.00018
Lange T, Samatov TR, Tonevitsky AG, Schumacher U. Importance of altered glycoprotein-bound N-and O-glycans for epithelial-to-mesenchymal transition and adhesion of cancer cells. Carbohydr. Res. 2014;389:39-45. https://doi.org/10.1016/j.carres.2014.01.010
Schwartz SL, Park EN, Vachon VK, Danzy S, Lowen AC, Conn GL. Human OAS1 activation is highly dependent on both RNA sequence and context of activating RNA motifs. Nucleic Acids Res. 2020;48(13):7520-7531. https://doi.org/10.1093/nar/gkaa513
Lohöfener J, Steinke N, Kay-Fedorov P, Baruch P, Nikulin A, Tishchenko S, Manstein DJ, Fedorov R. The activation mechanism of 2’-5’-oligoadenylate synthetase gives new insights into OAS/cGAS triggers of innate immunity. Structure. 2015;23(5):851-862. https://doi.org/10.1016/j.str.2015.03.012
Chen F, Wang Q, Zhou Y. The construction and validation of an RNA binding protein-related prognostic model for bladder cancer. BMC Cancer. 2021;21(1):244. https://doi.org/10.1186/s12885-021-07930-5
Wen X, Shao Z, Chen S, Wang W, Wang Y, Jiang J, Ma Q, Zhang L. Construction of an RNA-binding protein-related prognostic model for pancreatic adenocarcinoma based on TCGA and GTEx databases. Front. Genet. 2021;11:610350. https://doi.org/10.3389/fgene.2020.610350
Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta. 2013;1829(6-7):666-679. https://doi.org/10.1016/j.bbagrm.2013.02.003
Bakheet T, Hitti E, Khabar KSA. ARED-Plus: an updated and expanded database of AU-rich element-containing mRNAs and pre-mRNAs. Nucleic Acids Res. 2018;46(D1):D218-D220. https://doi.org/10.1093/nar/gkx975
Chan CP, Jin DY. Cytoplasmic RNA sensors and their interplay with RNA-binding partners in innate antiviral response: theme and variations. RNA. 2022;28(4):449-477. https://doi.org/10.1261/rna.079016.121
Chen M, Li S, Zhang J, Han J, Wang L, Yin D, Yi C, Yi Y. Exploration and verification of a six-RNA binding proteins-based prognosis evaluating model for hepatocellular carcinoma. November 2020. https://doi.org/10.21203/rs.3.rs-117829/v1
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Kletochnye Tekhnologii v Biologii i Meditsine, No. 1, pp. 34-39, March, 2023
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Novosad, V.O., Maltseva, D.V. The RNA-Binding Proteins OAS1, ZFP36L2, and DHX58 Are Involved in the Regulation of CD44 mRNA Splicing in Colorectal Cancer Cells. Bull Exp Biol Med 175, 144–149 (2023). https://doi.org/10.1007/s10517-023-05826-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10517-023-05826-x