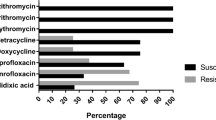
The tendency to the formation of macrolide resistance in campylobacteriosis pathogens is considered as a serious threat to public health due to ubiquity of campylobacter strains resistant to a wide range of antibiotics, primarily fluoroquinolones and tetracyclines. To assess the prevalence of resistant Campylobacter spp., we performed screening for macrolide sensitivity among 40 Campylobacter jejuni strains isolated from raw milk, poultry product, and washings from the equipment of the poultry processing plants. Phenotypic resistance to erythromycin, the most popular antibiotic for the treatment of campylobacteriosis, was revealed in 27.5% C. jejuni strains; 10% strains were resistant to azithromycin. The search and selection for gene markers of Campylobacter resistance to macrolides was performed. It was found that the resistance of C. jejuni to erythromycin is realized mainly via synthesis of proteins that protect ribosomes (the presence of coding sequences was detected in 45% of the studied strains) and the transmembrane pump mechanism (efflux pump CmeABC genes were found in 36% isolates); both mechanisms are transmissible. Chromosomal mutations in the 23S rRNA sequence detected in 18% strains seem to play a less significant role.
Similar content being viewed by others
References
Vinogradova KA, Bulgakova VG, Polin AN, Kozhevin PA. Microbial Antibiotic Resistance: Resistome, Its Volume, Diversity and Development. Antibiotiki Khimioterapiya. 2013;58(5-6):38-48. Russian.
Stetsenko VV, Efimochkina NR. The Mechanisms of Antibiotic Resistance in Bacteria of The Genus Campylobacter. Antibio-tiki Khimioterapiya. 2018;63(9-10):61-68. Russian.
Aminov RI, Mackie RI. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007;271(2):147-161.
Corcoran D, Quinn T, Cotter L, Fanning S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int. J. Antimicrob. Agents. 2006;27(1):40-45.
European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0, valid from 2015-01-01. URL: https://dvgmu.ru/images/data/pages/298/Kfz8MMFkDpU0s3Tf.pdf
Gibreel A, Kos VN, Keelan M, Trieber CA, Levesque S, Michaud S, Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicro. Agents Chemother. 2005;49(7):2753-2759.
Hao H, Yuan Z, Shen Z, Han J, Sahin O, Liu P, Zhang Q. Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 2013;57(3):1369-1378.
Iovine NM. Resistance mechanisms in Campylobacter jejuni. Virulence. 2013;4(3):230-240.
Ladely SR, Meinersmann RJ, Englen MD, Fedorka-Cray PJ, Harrison MA. 23S rRNA gene mutations contributing to macrolide resistance in Campylobacter jejuni and Campylobacter coli. Foodborne Pathog. Dis. 2009;6(1):91-98.
Ma L, Shen Z, Naren G, Li H, Xia X, Wu C, Shen J, Zhang Q, Wang Y. Identification of a novel G2073A mutation in 23S rRNA in amphenicol-selected mutants of Campylobacter jejuni. PLoS One. 2014;9(4). ID e94503. doi: https://doi.org/10.1371/journal.pone.0094503
Monier JM, Demanèche S, Delmont TO, Mathieu A, Vogel TM, Simonet P. Metagenomic exploration of antibiotic resistance in soil. Curr. Opin. Microbiol. 2011;14(3):229-235.
Pumbwe L, Piddock LJ. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol. Lett. 2002;206(2):185-189.
The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015;13(12). ID 4329. doi:https://doi.org/10.2903/j.efsa.2015.4329
Vacher S, Ménard A, Bernard E, Mégraud F. PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp.. Antimicrob. Agents Chemother. 2003;47(3):1125-1128.
Wei B, Kang M. Molecular basis of macrolide resistance in Campylobacter strains isolated from poultry in South Korea. Biomed Res. Int. 2018. ID 4526576. doi: https://doi.org/10.1155/2018/452657
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Byulleten’ Eksperimental’noi Biologii i Meditsiny, Vol. 169, No. 3, pp. 324-329, March, 2020
Rights and permissions
About this article
Cite this article
Efimochkina, N.R., Stetsenko, V.V. & Sheveleva, S.A. Formation of the Resistance of Campylobacter jejuni to Macrolide Antibiotics. Bull Exp Biol Med 169, 351–356 (2020). https://doi.org/10.1007/s10517-020-04885-8
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10517-020-04885-8