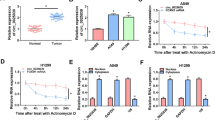
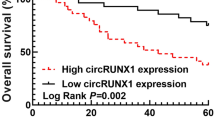
Abstract
Circular RNAs (circRNAs) are a specialized circular structure, are deregulated in cancers and play essential roles in biological processes involved in tumor progression. However, the mechanism by which circRNAs affect lung tumorigenesis and progression remains largely unexplored. To investigate the role of circRNA in lung cancer, circRNA expression profile was screened by bioinformatics analysis. The levels of circTAB2, miR-3142, and GLIS family zinc finger 2 (GLIS2) were measured by quantitate real-time (qRT-PCR) or western blot. Cell proliferation, apoptosis, migration and invasion were detected by EdU, flow cytometry, and transwell assays, respectively. Bioinformatics, western blot, RIP, pull down, dual luciferase reporter and rescue experiments were used to verify the direct relationship between miR-3142 and circTAB2 or GLIS2. The xenograft assays were used to assess the role of circTAB2 in vivo.CircTAB2 exhibited low expression in cancer tissues. Gain and loss-of-function assays indicated that circTAB2 could inhibit cell proliferation, migration and invasion. Functional studies revealed that circTAB2 acted as a miRNA sponge, directly interacted with miR-3142 and consequently regulated GLIS2 /AKT. Taken together, circTAB2 serves as an inhibitory role in lung cancer through a novel circTAB2 /miR-3142 /GLIS2 /AKT pathway and could be exploited a novel marker in lung cancer.






Similar content being viewed by others
Data Availability
The data and study materials that support the findings of this study will be available to other researchers from the corresponding authors on reasonable request.
References
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71:209–249
Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553:446–454
Li J, Kwok HF (2020) Current strategies for treating NSCLC: from biological mechanisms to clinical treatment. Cancers (Basel). https://doi.org/10.3390/cancers12061587
Jeck WR, Sorrentino JA, Wang K et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157
Vo JN, Cieslik M, Zhang Y et al (2019) The Landscape of Circular RNA in Cancer. Cell 176(869–881):e813
Shang Q, Yang Z, Jia R et al (2019) The novel roles of circRNAs in human cancer. Mol Cancer 18:6
Hua J, Wang X, Ma L et al (2022) CircVAPA promotes small cell lung cancer progression by modulating the miR-377-3p and miR-494-3p/IGF1R/AKT axis. Mol Cancer 21:123
Zhang LX, Gao J, Long X et al (2022) The circular RNA circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas via the miR-181a-5p/CARM1 axis. Mol Cancer 21:110
Wu D, Chen T, Zhao X et al (2022) HIF1alpha-SP1 interaction disrupts the circ-0001875/miR-31-5p/SP1 regulatory loop under a hypoxic microenvironment and promotes non-small cell lung cancer progression. J Exp Clin Cancer Res 41:156
Chen D, Zhou H, Cai Z et al (2022) CircSCAP interacts with SF3A3 to inhibit the malignance of non-small cell lung cancer by activating p53 signaling. J Exp Clin Cancer Res 41:120
Zhang T, Wu DM, Luo PW et al (2022) CircNEIL3 mediates pyroptosis to influence lung adenocarcinoma radiotherapy by upregulating PIF1 through miR-1184 inhibition. Cell Death Dis 13:167
Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21:22–36
Luo YH, Yang YP, Chien CS et al (2020) Plasma Level of Circular RNA hsa_circ_0000190 correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers (Basel) 12:1740
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20:1666–1670
Kent WJ, Sugnet CW, Furey TS et al (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Suzuki H, Zuo Y, Wang J et al (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34:e63
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Hauptmann J, Meister G (2013) Argonaute regulation: two roads to the same destination. Dev Cell 25:553–554
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48:D127–D131
Dudekula DB, Panda AC, Grammatikakis I et al (2016) CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13:34–42
Feng J, Chen W, Dong X et al (2022) CSCD2: an integrated interactional database of cancer-specific circular RNAs. Nucleic Acids Res 50:D1179–D1183
Sticht C, De La Torre C, Parveen A et al (2018) miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 13:e0206239
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
Szklarczyk D, Gable AL, Nastou KC et al (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612
Vivanco I, Sawyers CL (2002) The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2:489–501
Siegel RL, Miller KD, Fuchs HE et al (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33
Salmena L, Poliseno L, Tay Y et al (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Luo Q, Wang H, Li J (2019) Serum miR-3142 could be used as a potential biomarker to screen cervical cancer patients from healthy controls. Clin Lab. https://doi.org/10.7754/Clin.Lab.2018.180925
Zhang F, Jetten AM (2001) Genomic structure of the gene encoding the human GLI-related, Krüppel-like zinc finger protein GLIS2. Gene 280:49–57
Lichti-Kaiser K, ZeRuth G, Kang HS et al (2012) Gli-similar proteins: their mechanisms of action, physiological functions, and roles in disease. Vitam Horm 88:141–171
Wilson MM, Callens C, Le Gallo M et al (2021) An EMT-primary cilium-GLIS2 signaling axis regulates mammogenesis and claudin-low breast tumorigenesis. Sci Adv. https://doi.org/10.1126/sciadv.abf6063
Guerra E, Trerotola M, Aloisi AL et al (2013) The Trop-2 signalling network in cancer growth. Oncogene 32:1594–1600
Kim YS, Kang HS, Jetten AM (2007) The Krüppel-like zinc finger protein Glis2 functions as a negative modulator of the Wnt/beta-catenin signaling pathway. FEBS Lett 581:858–864
He L, Li Q, Du C et al (2022) Glis2 inhibits the epithelial-mesenchymal transition and apoptosis of renal tubule cells by regulating the beta-catenin signalling pathway in diabetic kidney disease. Biochem Biophys Res Commun 607:73–80
Sun H, Gu J, Li Z et al (2019) Low expression of GLIS2 gene might associate with radiosensitivity of gastric cancer. J Oncol 2019:2934925
Yao J, Lei PJ, Li QL et al (2020) GLIS2 promotes colorectal cancer through repressing enhancer activation. Oncogenesis 9:57
Funding
This work was supported by National Key Research and Development Program (2022YFC3600100, 2019YFA0903802) to YW, high-level talents of Youjiang Medical College for Nationalities (YY2021SK02) to LJ.
Author information
Authors and Affiliations
Contributions
Youliang Wang, Lihe Jiang and Yanli Lin designed and supervised the manuscript. Weiling Man, Yumeng Cui, Yanghua Li, Jie Jin, Yang Jin, Xiaojie Wu, Rongbin Zhong, Xiang Li and He Yao collected the data and conducted the statistical analyses. Weiling Man wrote the manuscript. Jie Li collected the clinical sample. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Ethical approval
Studies using human tissues and plasma samples were approved by the Ethics Committee of Chinese PLA General Hospital were performed in accordance with the principles of Declaration of Helsinki. The animal study was approved by the Animal Ethic Review Committees of Beijing Institute of Biotechnology. All animal experiments were strictly implemented in compliance with the NIH Guide for the Care and Use of Laboratory Animals.
Consent to participate and consent to publish
The patients provided their written informed consent to participate in this study. We have obtained consent to publish this paper from all the participants of this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.









Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Man, W., Cui, Y., Li, J. et al. circTAB2 inhibits lung cancer proliferation, migration and invasion by sponging miR-3142 to upregulate GLIS2. Apoptosis 28, 471–484 (2023). https://doi.org/10.1007/s10495-022-01805-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-022-01805-1