Abstract
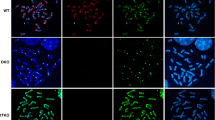
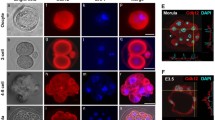
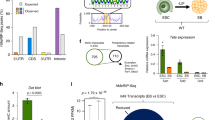
Retinoic acid-induced apoptosis of embryonic stem (ES) cells is an experimental system which resembles the physiological programmed cell death that occurs during differentiation in embryonic development. Our aim was to analyze the involvement of epigenetic modifications such as DNA methylation and chromatin structure in the apoptotic process and to investigate the metabolic activity of apoptotic bodies. We found a relationship between DNA methylation and apoptosis, shown by a dose-dependent induction of apoptosis after treatment with the inhibitor of DNA methylation 5-aza-2′-deoxycytidine. Interestingly, we found a slight demthylation of specific sequences of the U2afl-rs1 imprinted gene in those RA treated cells which were specifically undergoing apoptosis. In addition, apoptotic bodies exhibited an unexpected open chromatin conformation accessible to the endonuclease DNase-I. Furthermore, we observed a structural and functional preservation of specific DNA sequences and mRNA. These results suggest that biological activities, such as transcription or protein synthesis, could be maintained even towards the end of the apoptotic process.
Similar content being viewed by others
Abbreviations
- RA:
-
Retinoic acid
- LIF:
-
leukaemia inhibitory factor
- ES:
-
embryonic stem
- EC:
-
embryonal carcinoma
- AZC:
-
5-aza-cytidine
- AZDC:
-
5-aza-2′-deoxycytidine
- HSS:
-
hypersensitive site
- TSA:
-
trichostatin A.
References
Sluyser M (ed). Apoptosis in normal development and cancer. London: Taylor & Francis 1996
Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 1980; 284: 555–556
Häcker G. The morphology of apoptosis. Cell Tissue Res 2000; 301: 5–17
Robertson JD, Orrenius S, Zhivotovsky B. Review: Nuclear events in apoptosis. J Struct Biol 2000; 129: 346–358
Kerr JF, Wyllie AH, Currie AR. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 1972; 26: 239–257
Razin A, Cedar H. DNA methylation and genomic imprinting. Cell 1994; 77: 473–476
Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet 1997; 31: 493–525
Tilghman SM. The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell 1999; 96: 185–193
Jones PA. Cancer. Death and methylation. Nature 2001; 409: 141, 143–144
Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol 2002; 196: 1–7
Pompeia C, Hodge DR, Plass C, et al. Microarray analysis of epigenetic silencing of gene expression in the KAS-6/1 multiple myeloma cell line. Cancer Res 2004; 64: 3465–3473
Vanhaecke T, Papeleu, P Elaut G, Rogiers V. Trichostatin A-like hydroxamate histone deacetylase inhibitors as therapeutic agents: Toxicological point of view. Curr Med Chem 2004; 11: 1629–1643
Feil R, Khosla S. Genomic imprinting in mammals: An interplay between chromatin and DNA methylation? Trends Genet 1999; 15: 431–435
Bird A. The essentials of DNA methylation. Cell 1992; 70: 5–8
Cunliffe VT. Memory by modification: The influence of chromatin structure on gene expression during vertebrate development. Gene 2003; 305: 141–150
Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 1998; 19: 187–191
Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998; 393: 386–389
Drewell RA, Goddard CJ, Thomas JO, Surani MA. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res 2002; 30: 1139– 1144
Atencia R, Garcia-Sanz M, Unda F, Arechaga J. Apoptosis during retinoic acid-induced differentiation of F9 embryonal carcinoma cells. Exp Cell Res 1994; 214: 663–667
Asumendi A, Andollo N, Boyano MD, et al. The role of cleavage of cell structures during apoptosis. Cell Mol Biol 2000; 46: 1–11
Feil R, Boyano MD, Allen ND, Kelsey G. Parental chromosome-specific chromatin conformation in the imprinted U2af1-rs1 gene in the mouse. J Biol Chem 1997; 272: 20893–20900
Nabetani A, Hatada I, Morisaki H, Oshimura M, Mukai T. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol Cell Biol 1997; 17: 789–798
Hatada I, Sugama T, Mukai T. A new imprinted gene cloned by a methylation-sensitive genome scanning method. Nucleic Acids Res 1993; 21: 5577–5582
Hayashizaki Y, Shibata H, Hirotsune S, et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet 1994; 6: 33–40
Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 1994; 120: 2933–2943
Feil R, Handel MA, Allen ND, Reik W. Chromatin structure and imprinting: Developmental control of DNase-I sensitivity in the mouse insulin-like growth factor 2 gene. Dev Genet 1995; 17: 240–252
Oberhammer F, Wilson JW, Dive C, et al. Apoptotic death in epithelial cells: Cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. Embo J 1993; 12: 3679–3684
Tomei LD, Shapiro JP, Cope FO. Apoptosis in C3H/10T1/2 mouse embryonic cells: Evidence for internucleosomal DNA modification in the absence of double-strand cleavage. Proc Natl Acad Sci USA 1993; 90: 853–857
Wyllie A. Apoptosis. An endonuclease at last. Nature 1998; 391: 20–21
Saitoh F, Hiraishi K, Adachi M, Hozumi M. Induction by 5-aza-2′-deoxycytidine, an inhibitor of DNA methylation, of Le(y) antigen, apoptosis and differentiation in human lung cancer cells. Anticancer Res 1995; 15: 2137–2143
Jackson-Grusby L, Beard C, Possemato R, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 2001; 27: 31–39
Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. Embo J 2001; 20: 1963–1973
Gregory RI, O’Neill LP, Randall TE, et al. Inhibition of histone deacetylases alters allelic chromatin conformation at the imprinted U2af1-rs1 locus in mouse embryonic stem cells. J Biol Chem 2002; 277: 11728–11734
Widschwendter M, Berger J, Muller HM, Zeimet AG, Marth C. Epigenetic downregulation of the retinoic acid receptor-beta2 gene in breast cancer. J. Mammary Gland Biol. Neoplasia 2001; 6: 193–201
Berger J, Daxenbichler G. DNA methylation of nuclear receptor genes–possible role in malignancy. J. Steroid Biochem. Mol Biol 2002; 80: 1–11
Soengas MS, Capodieci P, Polsky D, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 2001; 409: 207–211
Fu WN, Bertoni F, Kelsey SM, et al. Role of DNA methylation in the suppression of Apaf–1 protein in human leukaemia. Oncogene 2003; 22: 451–455
Jablonka E, Goitein R, Marcus M, Cedar H. DNA hypomethylation causes an increase in DNase-I sensitivity and an advance in the time of replication of the entire inactive X chromosome. Chromosoma 1985; 93: 152–156
Lagarkova MA, Iarovaia OV, Razin SV. Large-scale fragmentation of mammalian DNA in the course of apoptosis proceeds via excision of chromosomal DNA loops and their oligomers. J Biol Chem 1995; 270: 20239–20241
Hendzel MJ, Nishioka WK, Raymond Y, Allis CD, Bazett-Jones DP, Th’ng JP. Chromatin condensation is not associated with apoptosis. J Biol Chem 1998; 273: 24470–24478
Kutsyi MP, Kuznetsova EA, Gaziev AI. Involvement of proteases in apoptosis. Biochemistry (Mosc) 1999; 64: 115– 126
Vizirianakis IS, Pappas IS, Gougoumas D, Tsiftsoglou AS. Expression of ribosomal protein S5 cloned gene during differentiation and apoptosis in murine erythroleukemia (MEL) cells. Oncol Res 1999; 11: 409–419
Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem 2001; 47: 1488– 1489
Lefresne J, Lemaitre JM, Selo M, Goussard J, Mouton C, Andeol Y. Evidence for multiple sequences and factors involved in c-myc RNA stability during amphibian oogenesis. Dev Growth Differ 2001; 43: 195–211
Atencia R, García-Sanz M, Pérez-Yarza G, Asumendi A, Hilario E, Aréchaga J. A structural analysis of cytoskeleton components during the execution phase of apoptosis. Protoplasma 1997; 198: 163–169
Atencia R, Asumendi A, Garcia-Sanz, M. Role of cytoskeleton in apoptosis. Vitam Horm 2000; 58: 267–297
Eguchi Y, Shimizu S, Tsujimoto Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res 1997; 57: 1835–1840
Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: A switch in the decision between apoptosis and necrosis. J Exp Med 1997; 185: 1481–1486
Hirsch T, Marchetti P, Susin SA, et al. The apoptosis-necrosis paradox. Apoptogenic proteases activated after mitochondrial permeability transition determine the mode of cell death. Oncogene 1997; 15: 1573–1581
Kroemer G, Dallaporta B, Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol 1998; 60: 619–642
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Andollo, N., Boyano, M.D., Andrade, R. et al. Structural and functional preservation of specific sequences of DNA and mRNA in apoptotic bodies from ES cells. Apoptosis 10, 417–428 (2005). https://doi.org/10.1007/s10495-005-0815-5
Issue Date:
DOI: https://doi.org/10.1007/s10495-005-0815-5