Abstract
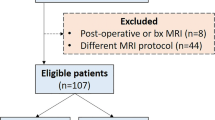
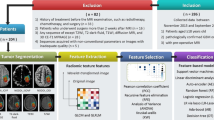
Texture-based convolutional neural networks (CNNs) have shown great promise in predicting various types of cancer, including lower grade glioma (LGG) through radiomics analysis. However, the use of CNN-based radiomics requires a large training set to avoid overfitting. To overcome this problem, the study proposes a novel panel of radiomic/texture features based on principal component analysis (PCA) applied to pretrained CNN features. The study used extracted PCA-CNN radiomic features from multimodal magnetic resonance imaging (MRI) images as input to a random forest (RF) classifier to predict immune cell markers, the gene status, and the survival outcome for LGG patients (n = 83). The results of the experiments demonstrate that RF with PCA-CNN radiomic features improved the classification performance, achieving the highest significant classification between short- and long-term survival outcomes. Notably, the area under the curve for PCA-CNN radiomic features with RF was 78.53% (p = 0.0008), which was significantly better than using gene status 63.14% (p = 0.23), clinical variables 52.60% (p = 0.32), standard radiomic features 72.56% (p = 0.02), immune cell markers 65.67% (p = 0.007), conditional entropy 74.54% (p = 0.0058), Gaussian mixture model-CNN 74.94% (p = 0.0053), or using 3D CNN classification directly without RF 72.61% (p = 0.01). The proposed PCA-CNN-based radiomic model outperformed state-of-the-art techniques to predict the survival outcome of LGG patients.







Similar content being viewed by others
Notes
References
Wang H, Hu J, Song Y, Zhang L, Bai S, Yi Z (2022) Multi-view fusion segmentation for brain glioma on ct images. Appl Intell 52(7):7890–7904
Mzoughi H, Njeh I, Wali A, Slima MB, BenHamida A, Mhiri C, Mahfoudhe KB (2020) Deep multi-scale 3d convolutional neural network (cnn) for mri gliomas brain tumor classification. J Digit Imaging 33:903–915
Su R, Liu X, Jin Q, Liu X, Wei L (2021) Identification of glioblastoma molecular subtype and prognosis based on deep mri features. Knowl-Based Syst 232:107490
Yin Q, Chen W, Zhang C, Wei Z (2022) A convolutional neural network model for survival prediction based on prognosis-related cascaded wx feature selection. Lab Invest 102(10):1064–1074
Goodenberger ML, Jenkins RB (2012) Genetics of adult glioma. Cancer Genet 205(12):613–621
Tom MC, Cahill DP, Buckner JC, Dietrich J, Parsons MW, Yu JS (2019) Management for different glioma subtypes: are all low-grade gliomas created equal? Am Soc Clin Oncol Educ Book 39:133–145
Chaichana KL, McGirt MJ, Laterra J, Olivi A, Quiñones-Hinojosa A (2010) Recurrence and malignant degeneration after resection of adult hemispheric low-grade gliomas. J Neurosurg 112(1):10–17
Hirata E, Sahai E (2017) Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med 7(7):026781
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ et al (2009) Idh1 and idh2 mutations in gliomas. N Engl J Med 360(8):765–773
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng H, Pfister SM, Reifenberger G et al (2021) The 2021 who classification of tumors of the central nervous system: a summary. Neurooncology 23(8):1231–1251
Liu X-Y, Gerges N, Korshunov A, Sabha N, Khuong-Quang D-A, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J et al (2012) Frequent atrx mutations and loss of expression in adult diffuse astrocytic tumors carrying idh1/idh2 and tp53 mutations. Acta Neuropathol 124:615–625
Network CGAR (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498
Lin W, Qiu X, Sun P, Ye Y, Huang Q, Kong L, Lu JJ (2021) Association of idh mutation and 1p19q co-deletion with tumor immune microenvironment in lower-grade glioma. Mol Ther Oncolytics 21:288–302
Wu F, Wang Z-L, Wang K-Y, Li G-Z, Chai R-C, Liu Y-Q, Jiang H-Y, Zhai Y, Feng Y-M, Zhao Z et al (2020) Classification of diffuse lowergrade glioma based on immunological profiling. Mol Oncol 14(9):2081
Yin W, Jiang X, Tan J, Xin Z, Zhou Q, Zhan C, Fu X, Wu Z, Guo Y, Jiang Z et al (2020) Development and validation of a tumor mutation burden–related immune prognostic model for lower-grade glioma. Front Oncol 10:1409
Fecci PE, Sampson JH (2019) The current state of immunotherapy for gliomas: an eye toward the future: Jnspg 75th anniversary invited review article. J Neurosurg 131(3):657–666
Liu Z, Wang S, Dong D, Wei J, Fang C, Zhou X, Sun K, Li L, Li B, Wang M et al (2019) The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 9(5):1303
Bhandari A, Liong R, Koppen J, Murthy S, Lasocki A (2021) Noninvasive determination of idh and 1p19q status of lower-grade gliomas using mri radiomics: A systematic review. Am J Neuroradiol 42(1):94–101
Shboul ZA, Chen J, Iftekharuddin KM (2020) Prediction of molecular mutations in diffuse low-grade gliomas using mr imaging features. Sci Rep 10(1):1–13
Cao M, Suo S, Zhang X, Wang X, Xu J, Yang W, Zhou Y (2021) Qualitative and quantitative MRI analysis in IDH1 genotype prediction of lower-grade gliomas: a machine learning approach. BioMed Res Int 2021:1235314. https://doi.org/10.1155/2021/1235314
Liu X-P, Jin X, SeyedAhmadian S, Yang X, Tian S-F, Cai Y-X, Chawla K, Snijders AM, Xia Y, van Diest PJ et al (2023) Clinical significance and molecular annotation of cellular morphometric subtypes in lower-grade gliomas discovered by machine learning. Neuro Oncol 25(1):68–81
Tang Z, Xu Y, Jin L, Aibaidula A, Lu J, Jiao Z, Wu J, Zhang H, Shen D (2020) Deep learning of imaging phenotype and genotype for predicting overall survival time of glioblastoma patients. IEEE Trans Med Imaging 39(6):2100–2109. https://doi.org/10.1109/TMI.2020.2964310
Trebeschi S, Drago S, Birkbak N, Kurilova I, Călin A, Delli Pizzi A, Lalezari F, Lambregts D, Rohaan M, Parmar C et al (2019) Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Annals of Oncology 30(6), 998–1004
Li Z-Z, Liu P-F, An T-T, Yang H-C, Zhang W, Wang J-X (2021) Construction of a prognostic immune signature for lower grade glioma that can be recognized by mri radiomics features to predict survival in lgg patients. Transl Oncol 14(6):101065
Kim AR, Choi KS, Kim M-S, Kim K-M, Kang H, Kim S, Chowdhury T, Yu HJ, Lee CE, Lee JH et al (2021) Absolute quantification of tumor-infiltrating immune cells in high-grade glioma identifies prognostic and radiomics values. Cancer Immunol Immunother 70:1995–2008
Zlochower A, Chow DS, Chang P, Khatri D, Boockvar JA, Filippi CG (2020) Deep learning ai applications in the imaging of glioma. Top Magn Reson Imaging 29(2):115–200
Decuyper M, Bonte S, Deblaere K, Van Holen R (2021) Automated mri based pipeline for segmentation and prediction of grade, idh mutation and 1p19q codeletion in glioma. Comput Med Imaging Graph 88:101831
Li Z, Wang Y, Yu J, Guo Y, Cao W (2017) Deep learning based radiomics (dlr) and its usage in noninvasive idh1 prediction for low grade glioma. Sci Rep 7(1):1–11
Chen T, Chefd’Hotel C (2014) Deep learning based automatic immune cell detection for immunohistochemistry images. In: International workshop on machine learning in medical imaging. Springer, Cham, pp 17–24
Ayadi W, Elhamzi W, Charfi I, Atri M (2021) Deep cnn for brain tumor classification. Neural Process Lett 53(1):671–700
Wang W, Liang D, Chen Q, Iwamoto Y, Han X-H, Zhang Q, Hu H, Lin L, Chen Y-W (2020) Medical image classification using deep learning. In: Deep learning in healthcare: paradigms and applications. Springer, pp 33–51
Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, Santamaría J, Fadhel MA, Al-Amidie M, Farhan L (2021) Review of deep learning: Concepts, cnn architectures, challenges, applications, future directions. J Big Data 8:1–74
Chaddad A, Peng J, Xu J, Bouridane A (2023) Survey of explainable ai techniques in healthcare. Sensors 23(2):634
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6(1):60
Chaddad A, Toews M, Desrosiers C, Niazi T (2019) Deep radiomic analysis based on modeling information flow in convolutional neural networks. IEEE Access 7:97242–97252
Chaddad A, Sargos P, Desrosiers C (2020) Modeling texture in deep 3d cnn for survival analysis. IEEE J Biomed Health Inform 25(7):2454–2462
Ma J, Yuan Y (2019) Dimension reduction of image deep feature using pca. J Vis Commun Image Represent 63:102578
Cascianelli S, Bello-Cerezo R, Bianconi F, Fravolini ML, Belal M, Palumbo B, Kather JN (2018) Dimensionality reduction strategies for cnn-based classification of histopathological images. In: International Conference on Intelligent Interactive Multimedia Systems and Services. Springer, Cham, pp 21–30
Kwak T, Song A, Kim Y (2019) The impact of the pca dimensionality reduction for cnn based hyperspectral image classification. Korean J Remote Sens 35(6 1):959–971
Shirahata M, Oba S, Iwao-Koizumi K, Saito S, Ueno N, Oda M, Hashimoto N, Ishii S, Takahashi JA, Kato K (2009) Using gene expression profiling to identify a prognostic molecular spectrum in gliomas. Cancer Sci 100(1):165–172
Cao H, Erson-Omay EZ, Li X, Günel M, Moliterno J, Fulbright RK (2020) A quantitative model based on clinically relevant mri features differentiates lower grade gliomas and glioblastoma. Eur Radiol 30:3073–3082
Chen Q, Han B, Meng X, Duan C, Yang C, Wu Z, Magafurov D, Zhao S, Safin S, Jiang C et al (2019) Immunogenomic analysis reveals lgals1 contributes to the immune heterogeneity and immunosuppression in glioma. Int J Cancer 145(2):517–530
Chaddad A, Hassan L, Desrosiers C (2021) Deep radiomic analysis for predicting coronavirus disease 2019 in computerized tomography and x-ray images. IEEE Trans Neural Netw Learn Syst 33(1):3–11
Zhao B, Dong X, Guo Y, Jia X, Huang Y (2022) PCA dimensionality reduction method for image classification. Neural Process Lett 54:347–368
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Yang T-HO, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA et al (2018) The immune landscape of cancer. Immunity 48(4):812–830
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sanchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Shao W, Wang T, Huang Z, Han Z, Zhang J, Huang K (2021) Weakly supervised deep ordinal cox model for survival prediction from whole-slide pathological images. IEEE Trans Med Imaging 40(12):3739–3747
Xu Y, Jia Z, Wang L-B, Ai Y, Zhang F, Lai M, Eric I, Chang C (2017) Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics 18(1):1–17
Bianchini M, Scarselli F (2014) On the complexity of neural network classifiers: A comparison between shallow and deep architectures. IEEE Trans Neural Netw Learn Syst 25(8):1553–1565. https://doi.org/10.1109/TNNLS.2013.2293637
Haralick RM (1979) Statistical and structural approaches to texture. Proc IEEE 67(5):786–804
Amadasun M, King R (1989) Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 19(5):1264–1274
Thibault G, Fertil B, Navarro C, Pereira S, Cau P, Levy N, Sequeira J, Mari J-L (2013) Shape and texture indexes application to cell nuclei classification. Int J Pattern Recognit Artif Intell 27(01):1357002
Lewandowski Z, Beyenal H (2013) Fundamentals of biofilm research. CRC Press, Boca Raton
Amit Y, Geman D (1997) Shape quantization and recognition with randomized trees. Neural Comput 9(7):1545–1588
Zweig MH, Campbell G (1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39(4):561–577
de Mello FL, Wilkinson JM, Kadirkamanathan V (2021) Metaparametric neural networks for survival analysis. IEEE Trans Neural Netw Learn Syst 1–10:3119510 https://doi.org/10.1109/TNNLS.2021
D’Arrigo G, Leonardis D, Abd ElHafeez S, Fusaro M, Tripepi G, Roumeliotis S (2021) Methods to analyse time-to-event data: the kaplan-meier survival curve. Oxidative Med Cell Longev 2021:2290120. https://doi.org/10.1155/2021/2290120
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE, Las Vegas, NV, pp 770–778
Redmon J, Farhadi A (2018) Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE, Salt Lake City, UT, pp 8697–8710
Ebrahimi A, Luo S, Chiong R (2020) Introducing transfer learning to 3D ResNet-18 for alzheimer’s disease detection on mri images. In: 2020 35th International Conference on Image and Vision Computing New Zealand (IVCNZ). IEEE, Wellington, New Zealand, pp 1–6
Lipková J, Angelikopoulos P, Wu S, Alberts E, Wiestler B, Diehl C, Preibisch C, Pyka T, Combs SE, Hadjidoukas P, Van Leemput K, Koumoutsakos P, Lowengrub J, Menze B (2019) Personalized radiotherapy design for glioblastoma: Integrating mathematical tumor models, multimodal scans, and bayesian inference. IEEE Trans Med Imaging 38(8):1875–1884 https://doi.org/10.1109/TMI.2019.2902044
Chaddad A, Desrosiers C, Abdulkarim B, Niazi T (2019) Predicting the gene status and survival outcome of lower grade glioma patients with multimodal mri features. IEEE Access 7:75976–75984
Zhou H, Vallieres M, Bai HX, Su C, Tang H, Oldridge D, Zhang Z, Xiao B, Liao W, Tao Y et al (2017) Mri features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19(6):862–870
Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, Wang Y, Chen L, Mao Y (2017) Noninvasive idh1 mutation estimation based on a quantitative radiomics approach for grade ii glioma. Eur Radiol 27(8):3509–3522
Li Y, Qian Z, Xu K, Wang K, Fan X, Li S, Jiang T, Liu X, Wang Y (2018) Mri features predict p53 status in lower-grade gliomas via a machine-learning approach. NeuroImage Clin 17:306–311
Deng X, Lin D, Chen B, Zhang X, Xu X, Yang Z, Shen X, Yang L, Lu X, Sheng H et al (2019) Development and validation of an idh1-associated immune prognostic signature for diffuse lower-grade glioma. Front Oncol 9:1310
Zhang M, Wang X, Chen X, Zhang Q, Hong J (2020) Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front Genet 11:363
Chan AK, Mao Y, Ng H-K (2016) Tp53 and histone h3. 3 mutations in triplenegative lower-grade gliomas. N Engl J Med 375(22):2206–2208
Zhang X, Liu S, Zhao X, Shi X, Li J, Guo J, Niedermann G, Luo R, Zhang X (2020) Magnetic resonance imaging-based radiomic features for extrapolating infiltration levels of immune cells in lower-grade gliomas. Strahlentherapie Onkol 1–9
Park C, Han K, Kim H, Ahn S, Choi D, Park Y, Chang J, Kim S, Cha S, Lee S (2021) Mri features may predict molecular features of glioblastoma in isocitrate dehydrogenase wild-type lower grade gliomas. Am J Neuroradiol 42(3):448–456
Ploug T, Holm S (2020) The four dimensions of contestable ai diagnostics-a patientcentric approach to explainable ai. Artif Intell Med 101901
Kumar N, Sukavanam N (2020) Weakly supervised deep network for spatiotemporal localization and detection of human actions in wild conditions. Vis Comput 36(9):1809–1821
Zhao M, Jia Z, Gong D (2019) Improved two-dimensional quaternion principal component analysis. IEEE Access 7:79409–79417
Zhao M, Jia Z, Cai Y, Chen X, Gong D (2021) Advanced variations of two-dimensional principal component analysis for face recognition. Neurocomputing 452:653–664
Acknowledgements
This work was supported by National Natural Science Foundation grant number 82260360, the Guilin Innovation Platform and Talent Program (20222C264164) and the Guangxi Science and Technology Based and Talent Project (2022AC18004, 2022AC21040).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflicts of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chaddad, A., Hassan, L. & Katib, Y. A texture-based method for predicting molecular markers and survival outcome in lower grade glioma. Appl Intell 53, 24724–24738 (2023). https://doi.org/10.1007/s10489-023-04844-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-023-04844-6