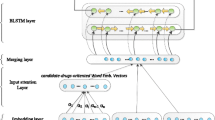
Abstract
Adverse drug reactions (ADRs), which are harmful physical reactions of patients to drug treatments, are inherent to the nature of drugs; the reactions can occur with any drug and are becoming a leading cause of patient morbidity and mortality during medical procedures. ADRs can be hazardous and even fatal to patients. In traditional methods, ADRs are detected through clinical trials. To obtain a comprehensive collection of ADRs, sufficient experimental samples and time are required before a drug comes to the market, which is not a realistic possibility. Moreover, even if extensive clinical trials are performed, many undetected ADRs might still be discovered after a drug is released to the market. ADRs can lead to disastrous consequences for humanity, which obviates a dramatically increased need for precise predictions of potential ADRs as early as possible. In this paper, we propose an encoder-decoder framework based on attention mechanism and the long short-term memory (LSTM) model to predict potential ADRs. We regard the prediction of ADRs as a sequence-to-sequence problem and improve the encoder-decoder framework based on the attention mechanism to learn the interrelationships between ADRs. Unlike other classical methods utilizing molecular drug structures, our model is based solely on ADRs, which is an independent but parallel approach compared to traditional methods. We capitalize on the mask method to generate the target data and use the 5-fold cross-validation method to cyclically verify the performance of our proposed model. Based on the Top-k accuracy test results, our model outperforms the baseline models in potential ADRs predictions.










Similar content being viewed by others
References
Coleman JJ, Pontefract SK (2016) Adverse drug reactions. Clin Med 16(5):48
Khalil H, Huang C (2020) Adverse drug reactions in primary care: a scoping review. BMC Health Serv Res 20(1):1–13
Edwards IR, Aronson JK (2000) Adverse drug reactions: definitions, diagnosis, and management. The lancet 356(9237):1255–1259
Marcum ZA, Handler SM, Boyce R, Gellad W, Hanlon JT (2010) Medication misadventures in the elderly: a year in review. Am J Geriatr Pharmacother 8(1):77–83
Schatz S, Weber RJ (2015) Adverse drug reactions Pharmacy Practice 1(1)
Hadi MA, Neoh CF, Zin RM, Elrggal ME, Cheema E (2017) Pharmacovigilance: pharmacists’ perspective on spontaneous adverse drug reaction reporting. Integr Pharm Res Pract 6:91
Bjarnason I (2013) Gastrointestinal safety of nsaids and over-the-counter analgesics. Int J Clin Pract 67:37–42
Zhang B, Xiong D, Su J (2018) Neural machine translation with deep attention. IEEE Trans Pattern Anal Mach Intell 42(1):154–163
Demner-Fushman D, Shooshan SE, Rodriguez L, Aronson AR, Lang F, Rogers W, Roberts K, Tonning J (2018) A dataset of 200 structured product labels annotated for adverse drug reactions. Scientific Data 5(1):1–8
Swathi T, Kasiviswanath N, Rao AA (2022) An optimal deep learning-based lstm for stock price prediction using twitter sentiment analysis. Applied Intelligence 1–14
Pauwels E, Stoven V, Yamanishi Y (2011) Predicting drug side-effect profiles: a chemical fragment-based approach. BMC bioinformatics 12(1):1–13
Liu M, Wu Y, Chen Y, Sun J, Zhao Z, Chen Xw, Matheny ME, Xu H (2012) Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc 19(e1):28–35
Huang LC, Wu X, Chen JY (2011) Predicting adverse side effects of drugs. BMC genomics 12(5):1–10
Zitnik M, Agrawal M, Leskovec J (2018) Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 34(13):457–466
Zhang W, Liu X, Chen Y, Wu W, Wang W, Li X (2018) Feature-derived graph regularized matrix factorization for predicting drug side effects. Neurocomputing 287:154–162
Deac A, Huang Y-H, Veličković P, Liò P, Tang J (2019) Drug-drug adverse effect prediction with graph co-attention CoRR arXiv:1905.00534
Xue R, Liao J, Shao X, Han K, Long J, Shao L, Ai N, Fan X (2019) Prediction of adverse drug reactions by combining biomedical tripartite network and graph representation model. Chem Res Toxicol 33(1):202–210
Ryu JY, Kim HU, Lee SY (2018) Deep learning improves prediction of drug–drug and drug–food interactions. Proc Natl Acad Sci 115(18):4304–4311
Zhang W, Yue X, Liu F, Chen Y, Tu S, Zhang X (2017) A unified frame of predicting side effects of drugs by using linear neighborhood similarity. BMC Syst Biol 11(6):23–34
Bean DM, Wu H, Iqbal E, Dzahini O, Ibrahim ZM, Broadbent M, Stewart R, Dobson RJ (2017) Knowledge graph prediction of unknown adverse drug reactions and validation in electronic health records. Sci Rep 7(1):1–11
Seo S, Lee T, Kim MH, Yoon Y (2020) Prediction of side effects using comprehensive similarity measures. BioMed research international 2020
Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, Bengio Y (2014) Learning phrase representations using rnn encoder-decoder for statistical machine translation Association for Computational Linguistics
Sutskever I, Vinyals O, Le QV (2014) Sequence to sequence learning with neural networks. In: Advances in neural information processing systems, pp 3104–3112
Bahdanau D, Cho K, Bengio Y (2015) Neural machine translation by jointly learning to align and translate. 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015 Conference Track Proceedings
Devlin J, Chang MW, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding Association for Computational Linguistics
Mullen LA, Benoit K, Keyes O, Selivanov D, Arnold J (2018) Fast, consistent tokenization of natural language text. J Open Source Softw 3(23):655
Yu Y, Si X, Hu C, Zhang J (2019) A review of recurrent neural networks: Lstm cells and network architectures. Neural Comput 31(7):1235–1270
Sherstinsky A (2020) Fundamentals of recurrent neural network (rnn) and long short-term memory (lstm) network. Phys D: Nonlinear Phenom 404:132306
Lamb AM, Goyal AGAP, Zhang Y, Zhang S, Courville AC, Bengio Y (2016) Professor forcing: A new algorithm for training recurrent networks. In: Advances in neural information processing systems, pp 4601–4609
Wu G, Liu J, Wang C (2017) Predicting drug-disease interactions by semi-supervised graph cut algorithm and three-layer data integration. BMC medical genomics 10(5):17–30
Jung Y (2018) Multiple predicting k-fold cross-validation for model selection. J Nonparametr Stat 30(1):197–215
Ji S, Pan S, Cambria E, Marttinen P, Philip SY (2021) A survey on knowledge graphs: Representation, acquisition, and applications. IEEE Transactions on Neural Networks and Learning Systems
Pereira DG, Afonso A, Medeiros FM (2015) Overview of friedman’s test and post-hoc analysis. Commun Stat Simul Comput 44(10):2636–2653
Probst P, Wright MN, Boulesteix AL (2019) Hyperparameters and tuning strategies for random forest. Wiley Interdiscip Rev Data Min Knowl Discov 9(3):1301
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No.62102241).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiahui Qian and Xihe Qiu contributed equally to this work
Rights and permissions
About this article
Cite this article
Qian, J., Qiu, X., Tan, X. et al. An Attentive LSTM based approach for adverse drug reactions prediction. Appl Intell 53, 4875–4889 (2023). https://doi.org/10.1007/s10489-022-03721-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-03721-y