Abstract
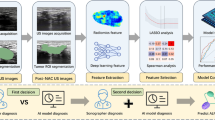
Among cancers, lung cancer has the highest morbidity, and mortality rate. The survival probability of lung cancer patients depends largely on an early diagnosis. For predicting lung cancer from low-dose Computed Tomography (LDCT) scans, computer-aided diagnosis (CAD) system needs to detect all pulmonary nodules, and combine their morphological features to assess the risk of cancer. An automatic lung cancer prognosis system is proposed. The existing CAD system is only for nodule detection. Actually, presence of a nodule does not mean cancer. Depending on its morphological features, the risk that it eventually would develop into cancer, is different. The motivation of the work is to propose a complete lung cancer prognosis system. It consists of 2 cascaded modules: nodule detection module and cancer risk evaluation module. In nodule detection module, two object detection algorithms are ensembled to minimize missing detection, i.e., maximize recall performance. They are based on 3D convolutional neural network (3D-CNN), and our recently proposed model of recurrent neural network (RNN). As they extract features in completely different ways, we call them heterogeneous deep learning models. By ensembing them, we could achieve much better recall performance compared to individual detectors. In cancer risk evaluation module, 3D-CNN based models are trained to evaluate the grade of malady of morphological features of pulmonary nodules. It will also provide medically interpretable intermediate information. Finally, a regression model is trained to match the ground truth labels describing morbidity grade of the CT-Scan. In this work, 13 features from the highest risk nodule is used to evaluate the risk of lung cancer. We also identify the subset of structural and morphological features which are strongly related to grading decision, labelled by oncologist. The final system could obtain a low logloss of 0.408.
















Similar content being viewed by others
References
American Cancer Society (2015) Global Cancer Facts & Figures 3rd Edition, pp 21
Fontana R S, Sanderson D R, Woolner L B, Taylor W F, Miller W E, Muhm J R (1986) Lung cancer screening: the Mayo program. J Occup Med 28(8):746–750
Ellert J, Kreel L (1980) The role of computed tomography in the initial staging and subsequent management of the lymphomas. J Comput Assist Tomogr 4(3):368–391
Bach P B, Kelley M J, Tate R C, McCrory D C (2003) Screening for lung cancer: a review of the current literature. Chest 123(1):72–82
Aberle D R, Adams A M, Berg C D, Black W C, Clapp J D, Fagerstrom R M, Gareen I F, Gatsonis C, Marcus P M, Sicks J D (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365(5):395–409
Singh S, Gierada D S et al (2012) Reader variability in identifying pulmonary nodules on chest radiographs from the national lung screening trial. J Thorac Imaging 27(4):249
Yu K H, Zhang C, Berry G J et al (2016) Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 7(1):1–10
Van Ginneken B, Setio A A A, Jacobs C et al (2015) Off-the-shelf Convolutional Neural Network Features for Pulmonary Nodule Detection in Computed Tomography Scans. In: Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp 286–289
Setio A A A, Ciompi F, Litjens G et al (2016) Pulmonary nodule detection in CT images: False positive reduction using Multi-View convolutional networks. IEEE Trans Med Imaging 35(5):1160–1169
Ding J, Li A, Hu Z, Wang L (2017) Accurate Pulmonary Nodule Detection in computed tomography images using deep convolutional neural networks. Medical Image Computing and Computer-Assisted Intervention, pp 559–567
Khosravan N, Bagci U (2017) S4ND: Single-shot Single-Scale lung nodule detection. Medical Image Computing and Computer-Assisted Intervention, pp 794–802
Dou Q, Chen H, Jin Y, Lin H, Qin J, Heng P A (2017) Automated Pulmonary Nodule Detection via 3D ConvNets with Online Sample Filtering and Hybrid-Loss Residual Learning. arXiv 2017, arXiv:1708.03867
Huang W, Xue Y, Wu Y (2019) A CAD system for pulmonary nodule prediction based on deep Three-Dimensional convolutional neural networks and ensemble learning. PLoS ONE 14(7):e0219369
LUNA16 Results. Available online: https://luna16.grand-challenge.org/Results/https://luna16.grand-challenge.org/Results/ (accessed on 7 January 2020)
Ypsilantis P, Montana G (2016) Recurrent convolutional networks for pulmonary nodule detection in CT imaging. arXiv:1609.09143
Riquelme D, Akhloufi M A (2020) Deep learning for lung cancer nodules detection and classification in CT scans. AI 1(1):28–67
Liao F, Liang M, Li Z et al (2019) Evaluate the Malignancy of Pulmonary Nodules Using the 3-D Deep Leaky Noisy-or Network. IEEE Trans Neural Netw Learn Syst 30(11):3484–3495
Wang W, Chakraborty G (2019) Evaluation of malignancy of lung nodules from CT image using recurrent neural network. IEEE international conference on systems, Man and Cybernetics:2992–2997
Wang W, Chakraborty G (2019) Deep Learning for Automatic Identification of Nodule Morphology Features and Prediction of Lung Cancer. 10th IEEE International Conference on Awareness Science and Technology, pp 1–6
Xu C, Hao K, Song Y (2013) Early diagnosis of solitary pulmonary nodules. J Thor Disease 5(6):830
Swensen S J, Silverstein M D, Ilstrup D M et al (1997) The probability of malignancy in solitary pulmonary nodules: application to small radiologically indeterminate nodules. Arch Internal Med 157(8):849–855
Lindell R M, Hartman T E, Swensen S J et al (2007) Five-year lung cancer screening experience: CT appearance, growth rate, location, and histologic features of 61 lung cancers. Radiol 2007 242(2):555–562
Daniel H (2017) Forecasting Lung Cancer Diagnoses with Deep Learning, https://dhammack.github.io/kaggle-ndsb2017/
The Cancer Imaging Archive, https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI
Neubeck A, Van Gool L (2006) Efficient non-maximum suppression. 18th International Conference on Pattern Recognition (ICPR’06), vol 3, pp 850–855
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Convolutional Neural Networks for Visual Recognition (2018) cs231n.github.io. Retrieved
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics, pp 315–323
Bahdanau D, Cho K, Bengio Y (2014) Neural machine translation by jointly learning to align and translate. arXiv:1409.0473
Deng P, Wang H, Li T et al (2019) Linear discriminant analysis guided by unsupervised ensemble learning. Inf Sci 480(4):211–221
Chen T, Guestrin C (2016) Xgboost: a scalable tree boosting system. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, pp 785–794
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors certify that there is no conflict of interest with any individual/organization for the present work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, W., Charkborty, G. Automatic prognosis of lung cancer using heterogeneous deep learning models for nodule detection and eliciting its morphological features. Appl Intell 51, 2471–2484 (2021). https://doi.org/10.1007/s10489-020-01990-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-020-01990-z