Abstract
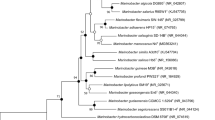
A novel Gram-staining-negative, rod-shaped, 0.6–0.8 µm wide and 2.0–3.0 µm in length, motile bacterium designated strain AK62T, was isolated from the green algal mat collected from saltpan, Kakinada, Andhra Pradesh, India. Colonies on ZMA were circular, off-white, shiny, moist, translucent, 1–2 mm in diameter, flat, with an entire margin. The major fatty acids include C16:0, C18:1 ω7c, and summed feature 3 (C16:1 ω7c and/or C16:1 ω6c and/or iso-C14:0 3-OH). Polar lipids include diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, one unidentified aminophospholipid, three unidentified phospholipids, and one unidentified lipid. Polyamine includes Spermidine. The DNA G + C content of the strain AK62T was 58.8 mol%. Phylogenetic analysis based on 16S rRNA gene sequence revealed that strain AK62T was closely related to the type strains Marinobacterium sediminicola, Marinobacterium coralli and Marinobacterium stanieri with a pair-wise sequence similarity of 96.9, 96.6 and 96.6%, respectively, forming a distinct branch within the genus Marinobacterium and clustered with M. stanieri, M. sediminicola, M. coralli and M. maritimum cluster. Strain AK62T shares average nucleotide identity (ANIb, based on BLAST) of 78.44, 76.69, and 76.95% with M. sediminicola CGMCC 1.7287T, M. stanieri DSM 7027T, and Marinobacterium halophilum Mano11T respectively. Based on the observed phenotypic, chemotaxonomic characteristics, and phylogenetic analysis, strain AK62T is described in this study as a novel species in the genus Marinobacterium, for which the name Marinobacterium alkalitolerans sp. nov. is proposed. The type strain of M. alkalitolerans is AK62T (= MTCC 12102T = JCM 31159T = KCTC 52667T).




Similar content being viewed by others
Data availability
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain AK62T is LN558833. The GenBank/EMBL/DDBJ accession number for the whole genome of strain AK62T is JACVEW000000000. All authors contributed equally to the work.
Availability of data and materials
The data and cultures are available.
Code availability
Here is a software application or custom code developed.
References
Alfaro-Espinoza G, Ullrich MS (2014) Marinobacterium mangrovicola sp. nov., a marine nitrogen-fixing bacterium isolated from mangrove roots of Rhizophora mangle. Int J Syst Evol Microbiol 64:3988–3993
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Baek SH, Cui Y, Kim SC, Cui CH, Yin C, Lee ST, Im WT (2011) Tumebacillus ginsengisoli sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol 61:1715–1719
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Baumann P, Bowditch RD, Baumann L, Beaman B (1983) Taxonomy of marine Pseudomonas species: P. stanieri sp. nov.; P. perfectomarina sp. nov., nom. Rev.; P. nautica; and P. doudoroffii. Int J Syst Bacteriol 33:857–865
Bernardet JF, Nakagawa Y, Holmes B (2002) Subcommittee on the taxonomy of Flavobacterium and Cytophaga–like bacteria of the International Committee on Systematics of Prokaryotes. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Chang HW, Nam YD, Kwon HY, Park JR, Lee JS, Yoon JH, An KG, Bae JW (2007) Marinobacterium halophilum sp. nov., a marine bacterium isolated from the Yellow Sea. Int J Syst Evol Microbiol 57:77–80
Chaudhari NM, Gupta VK, Dutta C (2016) BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373
Cowan ST, Steel KJ (1965) Manual for the Identification of Medical Bacteria, vol 149. Cambridge University Press, New York, p 852
González JM, Mayer F, Moran MA, Hodson RE, Whitman WB (1997) Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol 47:369–376
Gordon RE, Barnett DA, Handerhan JE, Pang CH-N (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Ivanova EP, Mikhaĭlov VV (2001) A new family of Alteromonadaceae fam. nov., including the marine proteobacteria species Alteromonas, Pseudoalteromonas, Idiomarina and Colwellia. Mikrobiologiia 70:15–23
Kim JM, Lee SH, Jung JY, Jeon CO (2010) Marinobacterium lutimaris sp. nov., isolated from a tidal flat. Int J Syst Evol Microbiol 60:1828–1831
Kim M, Oh H-S, Park S-C, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki K (1988) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Konstantinidis KT, Tiedje JM (2005) Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264
Lányí B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lee I, Chalita M, Ha SM, Na SI, Yoon SH, Chun J (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Sasser M (1990) Identification of bacteria through fatty acid analysis. In: Klement Z, Rudolph K, Sands Budapest DC (eds) Methods in phytobacteriology. Akademiai Kiado, Hungry, pp 199–204
Sly LI, Blackall LL, Kraat PC, Tao T-S, Sangkhobol V (1986) The use of second derivative plots for the determination of mol% guanine plus cytosine of DNA by the thermal denaturation method. J Microbiol Methods 5:139–156
Srinivas TNR, Bhumika V, Aditya S, Anil Kumar P (2014) Lunatimonas lonarensis gen. Nov., sp. Nov., a haloalkaline bacterium of the family Cyclobacteriaceae with nitrate reducing activity. Syst Appl Microbiol 37:10–16
Surendra V, Bhawana P, Suresh K, Srinivas TNR, Kumar PA (2012) Imtechella halotolerans gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from estuarine water. Int J Syst Evol Microbiol 62:2624–2630
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tanizawa Y, Fujisawa T, Nakamura Y (2018) DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34:1037–1039
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci USA 102:13950–13955
Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J (1996) Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60:407–438
Yarza P, Yilmaz P, Pruesse E et al (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. https://doi.org/10.1038/nrmicro3330
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu S, Zhu Z, Fu L, Niu B, Li W (2011) WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genom 12:444
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
The authors thank the Directors of, National Institute of Oceanography and Institute of Microbial Technology. We thank the Council of Scientific and Industrial Research (CSIR) and the Department of Biotechnology, Government of India, for financial assistance. TNRS is thankful for Projects OLP1710 and GAP 3195 for providing facilities and funding, respectively. We also thank Mr Deepak Bhatt for technical assistance in sequencing the 16S rRNA gene. GS thanks CSIR for the research fellowship. The NIO contribution number is 6725.
Funding
The work is supported by the funds received from the Institutional Project OLP1710 and Ministry of Earth Sciences Project GAP 3195.
Author information
Authors and Affiliations
Contributions
PAK, TNRS: Conceptualization, designed study, analyzed data, methodology, writing—original draft, Performed research: VG, GV, PSR, NK, SB, GSK.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pinnaka, A.K., Tanuku, N.R.S., Gupta, V. et al. Marinobacterium alkalitolerans sp. nov., with nitrate reductase and urease activity isolated from green algal mat collected from a solar saltern. Antonie van Leeuwenhoek 114, 1117–1130 (2021). https://doi.org/10.1007/s10482-021-01582-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01582-y