Abstract
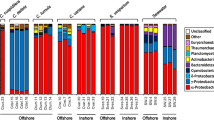
Marine sponges are abundant and ecologically important components of coral reefs and have been shown to harbour exceptionally high microbial densities, which can differ substantially among sponge species. However, this dichotomy between high and low microbial abundance (HMA, LMA) sponges is still not fully understood, particularly as concerns the archaeal community. This study aims to fill this gap by analysing (using 454-pyrosequencing of the 16S rRNA gene) how the archaeal community varies among known LMA (Stylissa carteri, and Stylissa massa), known HMA (Hyrtios erectus and Xestospongia testudinaria) and unknown HMA/LMA status sponge species (Ectyoplasia coccinea, Paratetilla bacca and Petrosia aff. spheroida) collected in a remote location in which very few sponge microbial composition studies have been previously performed (Mayotte, Comores archipelago, France) and comparing the results with those reported in four other geographical areas. Based on archaeal community composition, the known LMA sponges formed a distinct cluster together with Paratetilla bacca, Ectyoplasia coccinea and seawater while the known HMA sponge X. testudinaria formed a cluster with Petrosia aff. spheroida. The known HMA sponge H. erectus, in turn, had an intermediate archaeal community between HMA sponges and sediment samples. In addition to the above, we also showed significant compositional congruence between archaeal and bacterial communities sampled from the same sponge individuals. HMA sponges were mainly dominated by members assigned to the genus Nitrosopumilus while LMA sponges were mainly dominated by members assigned to the genus Cenarchaeum. In general, there was no clear difference in richness between HMA and LMA sponges. Evenness, however, was higher in HMA than LMA sponges. Whilst the present study corroborates some of the traits commonly associated with the HMA–LMA dichotomy (higher evenness in Mayotte HMA sponges), this was not consistent across geographical areas showing that more research is needed to fully understand the HMA/LMA dichotomy as concerns Archaea.





Similar content being viewed by others
Availability of data and materials
The DNA sequences generated in this study can be downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA): SRP071901: PRJNA315454.
References
Agnarsson I, Kuntner M (2012) The generation of a biodiversity hotspot: biogeography and phylogeography of the western Indian Ocean islands. Curr Topics Phylogenet Phylogeogr Terrest Aquat Syst 33:82
Alex A, Antunes A (2015) Pyrosequencing characterization of the microbiota from Atlantic intertidal marine sponges reveals high microbial diversity and the lack of co-occurrence patterns. PLoS ONE 10:e0127455
Bayer K, Schmitt S, Hentschel U (2008) Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol 10:2942–2955
Bayer K, Moitinho-Silva L, Brümmer F, Cannistraci CV, Ravasi T et al (2014a) GeoChip-based insights into the microbial functional gene repertoire of marine sponges (high microbial abundance, low microbial abundance) and seawater. FEMS Microbiol Ecol 90:832–843
Bayer K, Kamke J, Hentschel U (2014b) Quantification of bacterial and archaeal symbionts in high and low microbial abundance sponges using real-time PCR. FEMS Microbiol Ecol 89:679–690
Bell JJ (2008) The functional roles of marine sponges. Estuarine Coastal Shelf Sci 79:341–353. https://doi.org/10.1016/j.ecss.2008.05.002
Blanquer A, Uriz MJ, Galand PE (2013) Removing environmental sources of variation to gain insight on symbionts vs. transient microbes in high and low microbial abundance sponges. Environ Microbiol 15:3008–3019
Burton SA, Prosser JI (2001) Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol 67:2952–2957
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335
Chaib De Mares M, Sipkema D, Huang S, Bunk B, Overmann J, Van Elsas JD (2017) Host specificity for bacterial, archaeal and fungal communities determined for high-and low-microbial abundance sponge species in two genera. Front Microbiol 8:2560
Cleary DFR, de Voogd NJ, Polónia ARM, Freitas R, Gomes NCM (2015) Composition and predictive functional analysis of bacterial communities in seawater, sediment and sponges in an Indonesian coral reef environment. Microb Ecol 70:889–903. https://doi.org/10.1007/s00248-015-0632-5
Cleary DFR, Polónia ARM, Becking LE, de Voogd NJ, Purwanto GH, Gomes NCM (2018) Compositional analysis of bacterial communities in seawater, sediment and high and low microbial abundance sponges in the Misool coral reef system, Indonesia. Mar Biodivers 48:1889–1901. https://doi.org/10.1007/s12526-017-0697-0
Cleary DFR, Swierts T, Coelho FJ, Polónia ARM, Huang YM, Ferreira MR et al (2019) The sponge microbiome within the greater coral reef microbial metacommunity. Nat Commun 10:1644
Croué J, West NJ, Escande ML, Intertaglia L, Lebaron P, Suzuki MT (2013) A single betaproteobacterium dominates the microbial community of the crambescidine-containing sponge Crambe crambe. Sci Rep 3:2583
de Goeij JM, Van Oevelen D, Vermeij MJA, Osinga R, Middelburg JJ, de Goeij AFPM, Admiraal W (2013) Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342:108–110. https://doi.org/10.1126/science.1241981
de Voogd NJ, Cleary DFR, Polónia ARM, Gomes NC (2015) Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the thousand islands reef complex, West Java, Indonesia. FEMS Microbiol Ecol 91:fiv019
de Voogd NJ, Gauvin-Bialecki A, Polónia ARM, Cleary DFR (2019) Assessing the bacterial communities of sponges inhabiting the remote western Indian Ocean island of Mayotte. Mar Ecol 39:e12517
Diaz MC, Rützler K (2001) Sponges: an essential component of Caribbean coral reefs. Bull Mar Sci 69:535–546
Dupont S, Corre E, Li Y, Vacelet J, Bourguet-Kondracki ML (2013) First insights into the microbiome of a carnivorous sponge. FEMS Microbiol Ecol 86:520–531
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996
Edgar R, Haas B, Clemente J, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Erwin PM, Coma R, López-Sendino P, Serrano E, Ribes M (2015) Stable symbionts across the HMA–LMA dichotomy: low seasonal and interannual variation in sponge-associated bacteria from taxonomically diverse hosts. FEMS Microbiol Ecol 91:fiv115
Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, Thomas T (2012) Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA 109:E1878–E1887. https://doi.org/10.1073/pnas.1203287109
Francis CA, Beman JM, Kuypers MM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19
Gantt SE, McMurray SE, Stubler AD, Finelli CM, Pawlik JR, Erwin PM (2019) Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges. Microbiome 7:124
Giles EC, Kamke J, Moitinho-Silva L, Taylor MW, Hentschel U, Ravasi T, Schmitt S (2013) Bacterial community profiles in low microbial abundance sponges. FEMS Microbiol Ecol 83:232–241. https://doi.org/10.1111/j.1574-6941.2012.01467.x
Gloeckner V, Wehrl M, Moitinho-Silva L, Gernert C, Schupp P, Pawlik JR et al (2014) The HMA–LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol Bull 227:78–88
Gotelli NJ, Colwell RK (2001) Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett 4:379–391
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe YI, Sugahara J et al (2006a) Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA 103:18296–18301
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF (2006b) Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:e95
Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, Moore BS (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68:4431–4440
Hentschel U, Usher KM, Taylor MW (2006) Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55:167–177
Hentschel U, Piel J, Degnan SM, Taylor MW (2012) Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10:641–654. https://doi.org/10.1038/nrmicro2839
Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT, Schläppy M-L, Schleper C, Kuypers MMM (2009) Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11:2228–2243. https://doi.org/10.1111/j.1462-2920.2009.01944.x
Holmes B, Blanch H (2007) Genus-specific associations of marine sponges with Group I Crenarchaeotes. Mar Biol 150:759–772. https://doi.org/10.1007/s00227-006-0361-x
Jackson SA, Flemer B, McCann A, Kennedy J, Morrissey JP, O’Gara F, Dobson AD (2013) Archaea appear to dominate the microbiome of Inflatella pellicula deep sea sponges. PLoS ONE 8:e84438
Kamke J, Taylor MW, Schmitt S (2010) Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J 4:498
Kennedy J, Flemer B, Jackson SA, Morrissey JP, O’Gara F, Dobson AD (2014) Evidence of a putative deep sea specific microbiome in marine sponges. PLoS ONE 9:e91092
Kerou M, Schleper C (2015) Candidatus Cenarchaeum. Bergey's Manual of Systematics of Archaea and Bacteria, pp 1–4
Kiszka J, Ersts PJ, Ridoux V (2007) Cetacean diversity around the Mozambique Channel island of Mayotte (Comoros archipelago). J Cetacean Res Manag 9:105
Knobloch S, Jóhannsson R, Marteinsson V (2019) Bacterial diversity in the marine sponge Halichondria panicea from Icelandic waters and host-specificity of its dominant symbiont “Candidatus Halichondribacter symbioticus”. FEMS Microbiol Ecol 95:fiy220
Könneke M, Bernhard AE, José R, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543
Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, Qian PY (2011) Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J 5:650–664
Lenth R (2017) emmeans: estimated marginal means, aka least-squares means. https://CRAN.Rproject.org/package=emmeans
Lopes MF, Mágna B, Klautau M, Esteves EL, Albano RM (2019) Microbiota of the alien species Paraleucilla magna (Porifera, Calcarea) from the Southwestern Atlantic, and a comparison with that of other calcareous sponges. BioRxiv, 626192.
McMurray SE, Pawlik JR, Finelli CM (2014) Trait-mediated ecosystem impacts: how morphology and size affect pumping rates of the Caribbean giant barrel sponge. Aquat Biol 23:1–13. https://doi.org/10.3354/ab00612
Moitinho-Silva L, Seridi L, Ryu T, Voolstra CR, Ravasi T, Hentschel U (2014a) Revealing microbial functional activities in the Red Sea sponge Stylissa carteri by metatranscriptomics. Environ Microbiol 16:3683–3698
Moitinho-Silva L, Steinert G, Nielsen S, Hardoim CC, Wu YC, McCormack GP et al (2017) Predicting the HMA–LMA status in marine sponges by machine learning. Front Microbiol 8:752
Moitinho-Silva L, Bayer K, Cannistraci CV, Giles E, Ryu T, Seridi L et al (2014b) Specificity and transcriptional activity of microbiota associated with low and high microbial abundance sponges from the Red Sea. Mol Ecol 23:1348–1363
Oksanen J, Kindt R, Legendre P, O’Hara B, Simpson GL, Solymos P, Wagner H. 2009. Vegan: community ecology package. R Package Version. 1:15–14. http://www.cran.rproject.org/package=vegan
Pires AC, Cleary DFR, Almeida A, Cunha Â, Dealtry S, Mendonça-Hagler LC et al (2012) Denaturing gradient gel electrophoresis and barcoded pyrosequencing reveal unprecedented archaeal diversity in mangrove sediment and rhizosphere samples. Appl Environ Microbiol 78:5520–5528
Polónia ARM, Cleary DFR, Duarte LN, de Voogd NJ, Gomes NC (2014) Composition of Archaea in seawater, sediment, and sponges in the Kepulauan seribu reef system, Indonesia. Microb Ecol 67:553–567
Polónia ARM, Cleary DFR, Freitas R, de Voogd NJ, Gomes NC (2015) The putative functional ecology and distribution of archaeal communities in sponges, sediment and seawater in a coral reef environment. Mol Ecol 24:409–423
Polónia ARM, Cleary DFR, Freitas R, Coelho FJRC, de Voogd NJ, Gomes NCM (2016) Comparison of archaeal and bacterial communities in two sponge species and seawater from an Indonesian coral reef environment. Mar Genomics 29:69–80
Polónia ARM, Cleary DFR, Freitas R, Gomes NCM, de Voogd NJ (2017) Archaeal and bacterial communities of Xestospongia testudinaria and sediment differ in diversity, composition and predicted function in an Indonesian coral reef environment. J Sea Res 119:37–53
Polónia ARM, Cleary DFR, Coelho FJRC, Becking LE, de Voogd NJ, Toha AHA, Gomes NCM (2018) Compositional analysis of archaeal communities in high and low microbial abundance sponges in the Misool coral reef system, Indonesia. Mar Biol Res 14:537–550
Polónia ARM, Cleary DFR (2019) Archaeal communities in sponge, sediment and water from marine lakes and open water habitats. Mar Biol Res 15:259–274
Poppell E, Weisz J, Spicer L, Massaro A, Hill A, Hill M (2014) Sponge heterotrophic capacity and bacterial community structure in high-and low-microbial abundance sponges. Mar Ecol 35:414–424
Preston CM, Wu KY, Molinski TF, DeLong EF (1996) A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. P Nat Acad Sci 93:6241–6246
Qin W, Heal KR, Ramdasi R, Kobelt JN, Martens-Habbena W, Bertagnolli AD et al (2017) Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int J Syst Evol Microbiol 67:5067–5079
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C (2012) Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ Microbiol 14:909–923. https://doi.org/10.1111/j.1462-2920.2011.02661.x
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria ISBN 3-900051-07-0. http://www.R-project.orghttp://www.R-project.org
Said Hassane C, Fouillaud M, Le Goff G, Sklirou AD, Boyer JB, Trougakos IP et al (2020) Microorganisms associated with the marine sponge Scopalina hapalia: a reservoir of bioactive molecules to slow down the aging process. Microorganisms 8:1262
Schmitt S, Deines P, Behnam F, Wagner M, Taylor MW (2011) Chloroflexi bacteria are more diverse, abundant, and similar in high than in low microbial abundance sponges. FEMS Microbiol Ecol 78:497–510
Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008) Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74:7694–7708
Shannon E, Weaver W (1949) The Mathematical Theory of Communication. University of Illinois Press, Urbana
Thacker RW, Freeman CJ (2012) Sponge–microbe symbioses: recent advances and new directions. Adv Mar Biol 62:57–111
Thomas T, Rusch D, DeMaere MZ, Yung PY, Lewis M, Halpern A, Heidelberg KB, Egan S, Steinberg PD, Kjelleberg S (2010) Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J 4:1557–1567. https://doi.org/10.1038/ismej.2010.74
Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-Gárcia C, Olson JB, Erwin PM, Lopez-Legentil S, Luter H, Chaves-Fonnegra A, Costa R, Schupp PJ, Steindler L, Erpenbeck D, Gilbert J, Knight R, Ackermann G, Lopez JV, Taylor MW, Thacker RW, Montoya JM, Hentschel U, Webster NS (2016) Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7:1–12
Turon M, Uriz MJ (2020) New insights into the archaeal consortium of tropical sponges. Front Mar Sci 6:789. https://doi.org/10.3389/fmars.2019.00789
Turon M, Cáliz J, Garate L, Casamayor EO, Uriz MJ (2018) Showcasing the role of seawater in bacteria recruitment and microbiome stability in sponges. Sci Rep 8:1–10
Turque AS, Batista D, Silveira CB, Cardoso AM, Vieira RP, Moraes FC et al (2010) Environmental shaping of sponge associated archaeal communities. PLoS ONE 5:e15774
Tweedie MCK (1984) An index which distinguishes between some important exponential families. In: Ghosh JK, Roy J (eds) Statistics: applications and new directions—proceedings of the Indian Statistical Institute Golden Jubilee international conference. Indian Statistical Institute, Calcutta, pp 579–604
Weisz JB, Hentschel U, Lindquist N, Martens CS (2007) Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar Biol 152:475–483
Yahel G, Sharp JH, Marie D, Häse C, Genin A (2003) In situ feeding and element removal in the symbiont-bearing sponge Theonella swinhoei: bulk DOC is the major source for carbon. Limnol Oceanogr 48:141–149. https://doi.org/10.4319/lo.2003.48.1.0141
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Zhang F, Pita L, Erwin PM, Abaid S, López-Legentil S, Hill RT (2014) Symbiotic archaea in marine sponges show stability and host specificity in community structure and ammonia oxidation functionality. FEMS Microbiol Ecol 90:699–707. https://doi.org/10.1111/1574-6941.12427
Acknowledgements
Research permits were issued via the Prefecture of Mayotte. We thank Cécile Debitus, Bruno Fichou, Stephan Aubert, Philippe Prost, and Jean‐Pierre Bellanger for their support.
Funding
This study was financed through the ANR‐Netbiome under grant No ANR‐11‐EBIM‐0006 and a contribution to the project LESS CORAL [PTDC/AAC-AMB/115304/2009] funded by FEDER, through COMPETE2020 - Programa Operacional Competitividade e Internacionalização (POCI), and by national funds (OE), through the Portuguese Foundation for Science and Technology (FCT)/MCTES. Thanks are also due, to FCT/MCTES for the financial support to CESAM (UIDP/50017/2020 + UIDB/50017/2020), through national funds. Ana R.M. Polónia was supported by a postdoctoral scholarship (SFRH/BPD/117563/2016) funded by FCT/MCTES and by the European Social Fund (ESF).
Author information
Authors and Affiliations
Contributions
N.J.d.V. and D.F.R.C. designed the study; N.J.d.V. and A.G.B. collected the samples; A.R.M.P. performed the laboratory work; D.F.R.C. and A.R.M.P. performed the data analysis; A.R.M.P., D.F.R.C., A.G.B., and N.J.d.V wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors read and approved the final version of the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the supplementary information.
Supplementary Fig. 1
(a) Location map with (b) inset showing the island of Mayotte (PDF 182 kb)
Supplementary Fig. 2
Relative abundance of the most abundant classes in the demosponges Paratetilla bacca (Pb), Stylissa carteri (Sc), Stylissa massa (Sm), Ectyoplasia coccinea (Ec), Hyrtios erectus (He), Petrosia aff. spheroida (Ps), Xestospongia testudinaria (Xt); sediment (Sd), and seawater (Wt) (PDF 35 kb)
Supplementary Table 1
Sample list with the sample code, sponge species, Group code, Host, high microbial abundance (HMA) or low microbial abundance (LMA) type, collection site (location), and GPS coordinates (XLS 26 kb)
Supplementary Table 2
Results of emmeans analysis showing pairwise comparisons of differences in the relative abundances of selected phyla between biotopes based on the Tukey test. Significance: * 0.01 < Pr < 0.05 ** 0.001 < Pr < 0.01; *** Pr < 0.001 (XLS 52 kb)
Supplementary Table 3
Sample list with the sample code, biotope, number of replicates, total number of OTUs, number of core OTUs mean abundance of core OTUs, mean abundance of the single most abundant OTUs, number of specific OTUs, Diversity, Richness and Evenness. (XLS 36 kb)
Supplementary Table 4
Results of simper analysis showing the contribution of OTUs to differences in similarity between pairs of samples. OTUs that contribute significantly to differences are indicated: * 0.01 < P < 0.05 ** 0.001 < P < 0.01; *** P < 0.001 (XLSX 72 kb)
Supplementary Table 5
List of abundant (≥ 130 sequence reads) OTUs and closely related organisms identified using BLAST search. OTU: OTU number; Abund: number of sequence reads; Taxonomic classification, Acc: Genbank accession numbers of closely related organisms identified using BLAST; Seq: sequence similarity of these organisms with our representative OTU sequences; Source: isolation source of organisms identified using BLAST (XLS 36 kb)
Supplementary Table 6
List of abundant Mayotte OTUs with the same closely related organisms identified using BLAST search in other geographic areas. OTU: OTU number; Study: Geographic area; Reference: reference of the study in which the OTU were previous reported; Core: biotopes where the OTUs were core OTUs; Group: biotope where the OTUs were found; Abund: number of sequence reads; Database: Database used in QIIME; Taxonomic classification; GI:GenBank GenInfo sequence identifiers; Acc: Genbank accession numbers of closely related organisms identified using BLAST; Seq: sequence similarity of these organisms with our representative OTU sequences; Source: isolation source of organisms identified using BLAST; Location: Geographic region of the organisms identified using BLAST. In the ‘Group’ category, the most dominant OTUs and the OTUs predominantly found in a given biotope (not considering OTUs ≤ 5 sequences) are indicated by one asterisk (*) and/or marked bold respectively. Sm: S. massa; Sc: S. carteri; Wt: Water; He: H. erectus; Ps: Petrosia aff. spheroida; Sed: Sediment; Xt: X. testudinaria; Ap: Aaptos lobata; Bie: Biemna fortis. (XLS 41 kb)
Supplementary Table 7
Mean and standard deviation (SD) of Diversity (H′), Richness (S) and Evenness (J) indices reported in the LMA sponge species S. massa and S. carteri and the HMA sponge species X. testudinaria and H. erectus (N: number of replicates) across several geographic regions. Makassar: Spermonde Archipelago coral reef system; Jakarta: Kepulauan Seribu Reef System, Berau: Berau reef system and Papua: Misool coral reef system, Indonesia resorting to the results published in our previous studies (Polónia et al. 2014; 2015; 2016; 2017; 2018). (XLS 42 kb)
Supplementary Table 8
Mean and standard deviation (SD) relative abundance of the Thaumarchaeota and Euryarchaeota phyla reported in the LMA sponge species S. massa and S. carteri and the HMA sponge species X. testudinaria and H. erectus (N: number of replicates) across several geographic regions. Makassar: Spermonde Archipelago coral reef system; Jakarta: Kepulauan Seribu Reef System, Berau: Berau reef system and Papua: Misool coral reef system, Indonesia resorting to the results published in our previous studies (Polónia et al. 2014; 2015; 2016; 2017; 2018). (XLS 41 kb)
Supplementary Table 9
Number of core OTUs (C.OTUs) the mean abundance of these core OTUs (C.OTUs(%)) and the mean abundance of the single most abundant OTU (D.OTU(%)) in the LMA sponge species S. massa and S. carteri and the HMA sponge species X. testudinaria and H. erectus (N: number of replicates) across several geographic regions. Makassar: Spermonde Archipelago coral reef system; Jakarta: Kepulauan Seribu Reef System, Berau: Berau reef system and Papua: Misool coral reef system, Indonesia resorting to the results published in our previous studies (Polónia et al. 2014; 2015; 2016; 2017; 2018). (XLS 39 kb)
Rights and permissions
About this article
Cite this article
Polónia, A.R.M., Cleary, D.F.R., Gauvin‐Bialecki, A. et al. Archaeal communities of low and high microbial abundance sponges inhabiting the remote western Indian Ocean island of Mayotte. Antonie van Leeuwenhoek 114, 95–112 (2021). https://doi.org/10.1007/s10482-020-01503-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01503-5