Abstract
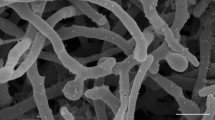
Two actinobacterial strains were isolated from samples collected from the University of Cape Town, South Africa. A third actinobacterial strain was isolated from soil collected in the town of Stellenbosch, South Africa, using a newly-developed Kribbella-selective medium. Analysis of the 16S rRNA genes showed that the three strains belonged to the genus Kribbella. A multilocus sequence analysis using the concatenated gene sequences of the gyrB, rpoB, relA, recA and atpD genes showed that strains YM55T and SK5 were most closely related to the type strains of Kribbella sindirgiensis and Kribbella soli, while strain YM53T was most closely related to the type strain of Kribbella pittospori. Digital DNA–DNA hybridisation and Average Nucleotide Identity (ANI) analyses showed that strains YM55T and SK5 belong to the same genomic species (OrthoANI value = 98.4%), but are distinct from the genomic species represented by the type strains of K. sindirgiensis (OrthoANI values < 95.6%) and K. soli (OrthoANI values < 91.4%). Strain YM53T is distinct from the genomic species represented by the type strain of K. pittospori (OrthoANI value = 94.0%). Phenotypic comparisons showed that strains YM55T and SK5 are distinct from the type strains of K. sindirgiensis and K. soli and that strain YM53T is distinct from the type strain of K. pittospori. Strains YM53T and YM55T are thus presented as the type strains of novel species, for which the names Kribbella capetownensis sp. nov. (= DSM 29426T = NRRL B-65062T) and Kribbella speibonae sp. nov. (= DSM 29425T = NRRL B-59161T), respectively, are proposed.



Similar content being viewed by others
References
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc)
Atlas RM (2004) Handbook of Microbiological Media, 3rd edn. CRC Press, Boca Raton
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Bertels F, Silander OK, Pachkov M, Rainey PB, Van Nimwegen E (2014) Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:81–87
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M (2018) Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392
Cook AE, Meyers PR (2003) Rapid identification of filamentous actinomycetes to the genus level using genus-specific 16S-rRNA gene restriction fragment patterns. Int J Syst Evol Microbiol 53:1907–1915
Curtis SM, Meyers PR (2012) Multilocus sequence analysis of the actinobacterial genus Kribbella. Syst Appl Microbiol 35:441–446
Curtis SM, Norton I, Everest GJ, Meyers PR (2018) Kribbella podocarpi sp. nov., isolated from the leaves of a yellowwood tree (Podocarpus latifolius). Antonie Van Leeuwenhoek 111:875–882
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Everest GJ, Cook AE, Kirby BM, Meyers PR (2011) Evaluation of the use of recN sequence analysis in the phylogeny of the genus Amycolatopsis. Antonie Van Leeuwenhoek 100:483–496
Everest GJ, Curtis SM, De Leo F, Urzì C, Meyers PR (2013) Kribbella albertanoniae sp. nov., isolated from a Roman catacomb, and emended description of the genus Kribbella. Int J Syst Evol Microbiol 63:3591–3596
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Kaewkla O, Franco CMM (2013) Kribbella endophytica sp. nov., an endophytic actinobacterium isolated from the surface-sterilized leaf of a native apricot tree. Int J Syst Evol Microbiol 63:1249–1253
Kaewkla O, Franco CM (2016) Kribbella pittospori sp. nov., an endophytic actinobacterium isolated from the surface-sterilized stem of an Australian native apricot tree, Pittosporum angustifolium. Int J Syst Evol Microbiol 66:2284–2290
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirby BM, Everest GJ, Meyers PR (2010) Phylogenetic analysis of the genus Kribbella based on the gyrB gene: proposal of a gyrB-sequence threshold for species delineation in the genus Kribbella. Antonie Van Leeuwenhoek 97:131–142
Komagata K, Suzuki KL (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee I, Kim YO, Park SC, Chun J (2015) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Li WJ, Wang D, Zhang YQ, Schumann P, Stackebrandt E, Xu LH, Jiang CL (2004) Kribbella antibiotica sp. nov., a novel nocardioform actinomycete strain isolated from soil in Yunnan, China. Syst Appl Microbiol 27:160–165
Matson JA, Bush JA (1989) Sandramycin, a novel antitumor antibiotic produced by a Nocardioides sp. Production, isolation, characterization and biological properties. J Antibiot (Tokyo) 42:1763–1767
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J Microbiol Methods 2:233–241
Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, Pukall R, Klenk H-P, Goodfellow M, Göker M (2018) Genome-Based Taxonomic Classification of the Phylum Actinobacteria. Front Microbiol 9:2007
Ozdemir-Kocak F, Saygin H, Saricaoglu S, Cetin D, Guven K, Spröer C, Schumann P, Klenk HP, Sahin N, Isik K (2017a) Kribbella soli sp. nov., isolated from soil. Antonie Van Leeuwenhoek 110:641–649
Ozdemir-Kocak F, Isik K, Saricaoglu S, Saygin H, Inan-Bektas K, Cetin D, Guven K, Sahin N (2017b) Kribbella sindirgiensis sp. nov. isolated from soil. Arch Microbiol 199:1399–1407
Park YH, Yoon JH, Shin YK, Suzuki K, Kudo T, Seino A, Kim H, Lee JS, Lee ST (1999) Classification of ‘Nocardioides fulvus’ IFO 14399 and Nocardioides sp. ATCC 39419 in Kribbella gen. nov., as Kribbella flavida sp. nov. and Kribbella sandramycini sp. nov. Int J Syst Bacteriol 49:743–752
Patrick S, McDowell A (2012) Order XII. Propionibacteriales ord. nov. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol 5, 2nd edn. Springer, New York, p 1137
Saygin H, Ay H, Guven K, Sahin N (2019) Kribbella turkmenica sp. nov., isolated from the Karakum Desert. Int J Syst Evol Microbiol 69:2533–2540
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Sohn K, Hong SG, Bae KS, Chun J (2003) Transfer of Hongia koreensis Lee et al. 2000 to the genus Kribbella Park et al. 1999 as Kribbella koreensis comb. nov. Int J Syst Evol Microbiol 53:1005–1007
Song W, Duan L, Zhao J, Jiang S, Guo X, Xiang W, Wang X (2018) Kribbella monticola sp. nov., a novel actinomycete isolated from soil. Int J Syst Evol Microbiol 68:3441–3446
Sun JQ, Xu L, Guo Y, Li WL, Shao ZQ, Yang YL, Wu XL (2017) Kribbella deserti sp. nov., isolated from rhizosphere soil of Ammopiptanthus mongolicus. Int J Syst Evol Microbiol 67:692–696
Trujillo ME, Kroppenstedt RM, Schumann P, Martinez-Molina E (2006) Kribbella lupini sp. nov., isolated from the roots of Lupinus angustifolius. Int J Syst Evol Microbiol 56:407–411
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Williams ST, Goodfellow M, Alderson G (1989) Genus Streptomyces Waksman and Henrici 1943, 339AL. In: Williams ST, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 4. Williams & Wilkins, Baltimore, pp 2452–2492
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhao J, Duan L, Qian L, Cao P, Tian Y, Ju H, Xiang W, Wang X (2019) Kribbella jiaozuonensis sp. nov., a novel actinomycete isolated from soil. Int J Syst Evol Microbiol 69:3500–3507
Acknowledgements
SMC held a National Research Foundation (NRF) Innovation Doctoral Scholarship. GJE held a Claude Leon Foundation Postdoctoral Fellowship. This work was supported by research grants to P.R.M. from the National Research Foundation (Grant Nos. 81014 and 85476) and the University Research Committee (University of Cape Town). Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the NRF.
Author information
Authors and Affiliations
Contributions
SMC Developed the Kribbella selective medium, wrote the original manuscript (ANTO-D-14-00425), did all the multilocus-sequence analyses (MLSA) and genetic-distance calculations and constructed the concatenated-gene phylogenetic tree. Proof-read the manuscript. IN Isolated strains YM53T and YM55T and carried out the initial characterisation of these strains. GJE Carried out the polar-lipid analyses of strains YM53T and YM55T. Proof-read the manuscript. JGP Carried out a full polyphasic characterisation of strain SK5 (as a possible new species) as part of his M.Sc. studies. Proof-read the manuscript. MCdK Isolated strain SK5, carried out the initial characterisation of the strain and determined the sequences of the gyrB, rpoB, relA, recA and atpD genes. Proof-read the manuscript. PRM Carried out the comparative phenotypic testing on strains YM53T, YM55T and SK5 and their revised closest phylogenetic relatives. Constructed the 16S rRNA gene phylogenetic tree and did the genomic and phylogenomic analyses. Re-wrote the 2014 manuscript to include the updated information. Supervised the research and provided the research funding.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Curtis, S.M., Norton, I., Everest, G.J. et al. Development of a Kribbella-specific isolation medium and description of Kribbella capetownensis sp. nov. and Kribbella speibonae sp. nov., isolated from soil. Antonie van Leeuwenhoek 113, 617–628 (2020). https://doi.org/10.1007/s10482-019-01365-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-019-01365-6