Abstract
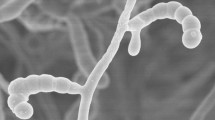
A novel actinomycete, designated strain XZ 46T, was isolated from acid sandy soil collected from the Tibetan Plateau, China. Its taxonomic position was determined using a polyphasic approach. Strain XZ 46T shows the typical morphological and chemotaxonomic features of members of the genus Streptomyces: slightly yellow to brown substrate mycelia and grayish white to slightly yellow aerial hyphae forming cylindrical and spiny spores; meso-diaminopimelic acid in the cell wall peptidoglycan; MK-9(H8), MK-9(H4) and MK-9(H2) as predominant menaquinones; diphosphatidylglycerol, phospatidylethanolamine, phosphatidylglycerol and phosphatidylinositol as main polar lipids; and iso-C15:0, iso-C16:0 and anteiso-C15:0 as major cellular fatty acids. The G+C content of the draft genome sequence, consisting of 8,995,813 bp, is 71.23%. The16S rRNA gene sequence analysis indicated that strain XZ 46T shows high sequence similarity to Streptomyces luteogriseus NBRC 13402T as well as forming an independent lineage clade with it in phylogenetic trees. Multilocus sequence analysis (MLSA) of five housekeeping genes (atpD, gyrB, recA, rpoB and trpB) illustrated that Streptomyces hawaiiensis is also a very closely related taxon. However, DNA–DNA hybridization, MLSA evolutionary distance and phenotypic properties demonstrate that strain XZ 46T can be distinguished from these phylogenetically related Streptomyces species. Therefore, it is concluded that strain XZ 46T represents a novel species of the genus Streptomyces, for which the name Streptomyces tibetensis is sp. nov. proposed. The type strain is XZ 46T (= CGMCC 4.7579T = KCTC 49221T).


Similar content being viewed by others
References
Collee JG, Miles RS, Watt B (1996) Tests for identification of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A (eds) Practical medical microbiology, 14th edn. Churchill Livingstone, New York, pp 131–149
Collins MD, Howarth OW, Grund E, Kroppenstedt RM (1987) Isolation and structural determination of new members of the vitamin K2 series in Nocardia brasiliensis. FEMS Microbiol Lett 41:35–39
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Sys Zool 20:406–416
Goodfellow M (1971) Numerical taxonomy of some nocardioform bacteria. J Gen Microbiol 69:33–80
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Hayakawa M (2008) Studies on the isolation and distribution of rare actinomycetes in soil. Actinomycetologica 22:12–19
Hsu SC, Lockwood JL (1975) Powdered chitin agar as selective medium for enumeration of actinomycetes in water and soil. Appl Microbial 29:422–426
Huss VA, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11:1–11
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol 42:989–1005
Kelly KL (1964) Inter-society color council-national bureau of standards color-name charts illustrated with centroid colors. US Government Printing Office, Washington, DC
Komagata K, Suzuki K (1988) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger data sets. Mol Biol Evol 33:1870–1874
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104
Labeda DP, Doroghazi JP, Ju K-S, Metcalf WW (2014) Taxonomic evaluation of Streptomyces albus and related species using multi-locus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int J Syst Evol Microbiol 64:894–900
Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, Ju KS, Metcalf WW (2017) Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek 110:563–583
Lechevalier MP, Lechevalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Evol Microbiol 20:435–443
Li Y, Li Y, Wang LW, Bao J (2018) Streptomyces dengpaensis sp. nov., an actinomycete isolated from desert soil. Int J Syst Evol Microbiol 68:3322–3326
Li L, Wang J, Zhou YJ, Lin HW, Lu YH (2019) Streptomyces reniochalinae sp. nov. and Streptomyces diacarni sp. nov., from marine sponges. Int J Syst Evol Microbiol 69:99–104
Liu R, Deng Z, Liu T (2018) Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab Eng 50:74–84
Rong X, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multi-locus sequence analysis and DNA–DNA hybridization, validating the MLSA scheme for the systematics of the whole genus. Syst Appl Microbiol 35:7–18
Röttig A, Atasayar E, Meier-Kolthoff JP, Spröer C, Schumann P, Schauer J, Steinbüchel A (2017) Streptomyces jeddahensis sp. nov., an oleaginous bacterium isolated from desert soil. Int J Syst Evol Microbiol 67(6):1676–1682
Saeng-In P, Phongsopitanun W, Savarajara A, Tanasupawat S (2018) Streptomyces lichenis sp. nov., isolated from lichen. Int J Syst Evol Microbiol 68:3641–3646
Saitou N, Nei M (1987) The neighbor joining method: a new method or reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc, Newark
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Shirling EB, Gottlieb D (1969) Cooperative description of type cultures of Streptomyces. IV. Species descriptions from the second, third and fourth studies. Int J Syst Bacteriol 19:391–512
Tadashi A (1975) Culture media for actinomycetes. The Society for Actinomycetes, Tokyo
Tindall BJ, Sikorski J, Smibert RM, Krieg NR (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, et al. (eds) Methods for general and molecular microbiology, 3rd edn. ASM Press, Washington, DC, pp 330–393
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:464
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:237–243
Weiburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Williams ST, Cross T (1971) Actinomycetes. Methods Microbiol 4:295–334
Williams ST, Goodfellow M, Alderson G, Wellington EM, Sneath PH, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang B, Tang S, Chen X, Zhang L, Zhang G, Zhang W, Liu G, Chen T, Li S, Dyson P (2016) Streptomyces lacrimifluminis sp. nov., a novel actinobacterium that produces antibacterial compounds, isolated from soil. Int J Syst Evol Microbiol 66:4981–4986
Zhang B, Tang S, Yang R, Chen X, Zhang D, Zhang W, Li S, Chen T, Liu G, Dyson P (2019) Streptomyces dangxiongensis sp. nov., isolated from soil of Qinghai-Tibet Plateau. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.003550
Acknowledgements
Thanks to National Natural Science Foundation of China (Grant No. U1806222), Shandong Provincial Natural Science Joint Found With Universities and Scientific Research Institution (ZR2018LC001).
Author information
Authors and Affiliations
Contributions
JL and LW conducted the experiments. ZY and LL contributed to writing. YL was in charge of experimental expenditure.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Wang, L., Ye, Z. et al. Streptomyces tibetensis sp. nov., an actinomycete isolated from the Tibetan Plateau. Antonie van Leeuwenhoek 113, 33–41 (2020). https://doi.org/10.1007/s10482-019-01315-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-019-01315-2