Abstract
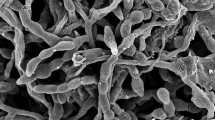
A novel actinobacterial strain, MKSP12T, was isolated from coastal sediment of a crater lake in central Anatolia, Turkey. The taxonomic position of the strain was clarified using a polyphasic approach. Phylogenetic analyses based on 16S rRNA gene sequences showed that strain MKSP12T is closely related to Streptomyces specialis GW 41-1564T with 97.1% sequence similarity. The strain produces aerial hyphae that differentiate into spiral chains of smooth surfaced spores and grows over a temperature range of 20–37 °C, at pH 7–11 and in the presence of 3% (w/v) sodium chloride. The cell wall amino acid is LL-diaminopimelic acid and the whole cell sugars are glucose and ribose. The polar lipids profile consists of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, an unidentified aminophospholipid, two unidentified phospholipids, an unidentified glycophospholipid and eight unidentified glycolipids; iso-C16:0, iso-C16:1 G, anteiso-C17:0 and anteiso-C17:1 ω9c were identified as the predominant cellular fatty acids (> 10%). Based on morphological and chemotaxonomic characteristics, and phylogenetic analyses, the strain is considered to represent a novel species in the genus Streptomyces, for which the name Streptomyces sediminis sp. nov. is proposed with the type strain MKSP12T (= DSM 100692T = KCTC 39613T).


Similar content being viewed by others
References
Akhwale JK, Göker M, Rohde M, Spröer C, Schumann P, Klenk H-P, Boga HI (2015) Streptomyces alkaliphilus sp. nov., isolated from sediments of Lake Elmenteita in the Kenyan Rift Valley. Antonie Van Leeuwenhoek 107:1249–1259
Baranasic D, Gacesa R, Starcevic A, Zucko J, Blažič M, Horvat M, Gjuračić K et al (2013) Draft genome sequence of Streptomyces rapamycinicus strain NRRL 5491, the producer of the immunosuppressant rapamycin. Genome Announc 1:e00513–e00581
Bian G-K, Qin S, Yuan B, Zhang Y-J, Xing K, Ju X-Y, Li W-J et al (2012) Streptomyces phytohabitans sp. nov., a novel endophytic actinomycete isolated from medicinal plant Curcuma phaeocaulis. Antonie Van Leeuwenhoek 102:289–296
Braña AF, Fiedler H-P, Nava H, González V, Sarmiento-Vizcaíno A, Molina A, Acuña JL et al (2015) Two Streptomyces species producing antibiotic, antitumor, and anti-inflammatory compounds are widespread among intertidal macroalgae and deep-sea coral reef invertebrates from the central cantabrian sea. Microb Ecol 69:512–524
Busarakam K, Bull AT, Girard G, Labeda DP, van Wezel GP, Goodfellow M (2014) Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie Van Leeuwenhoek 105:849–861
Chen HH, Qin S, Lee JC, Kim CJ, Xu LH, Li WJ (2009) Streptomyces mayteni sp.nov., a novel actinomycete isolated from a Chinese medicinal plant. Antonie Van Leeuwenhoek 95:47–53
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Evol Microbiol 45:240–245
de Lima Procópio RE, da Silva IR, Martins MK, de Azevedo JL, de Araújo JM (2012) Antibiotics produced by Streptomyces. Braz J Infect Dis 16:466–471
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Erener A, Yakar M (2012) Monitoring coastline change using remote sensing and GIS technologies. Lecture Notes Inf Technol 30:310–314
Et Shirling, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol 16:313–340
Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24:774–786
Goodfellow M, Fiedler HP (2010) A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98:119–142
Gregersen T (1978) Rapid method for distinction of Gram-negative from Gram-positive bacteria. Appl Microbiol Biotechnol 5:123–127
He H, Liu C, Zhao J, Li W, Pan T, Yang L, Wang X, Xiang W (2014) Streptomyces zhaozhouensis sp. nov., an actinomycete isolated from candelabra aloe (Aloe arborescens Mill). Int J Syst Evol Microbiol 64:1096–1101
Huang X, Zhou S, Huang D, Chen J, Zhu W (2015) Streptomyces spongiicola sp. nov., a novel marine sponge-derived actinomycete. Int J Syst Evol Microbiol 66:738–743
Ichimura M, Ogawa T, Katsumata S, K-i Takahashi, Takahashi I, Nakano H (1991) Duocarmycins, new antitumor antibiotics produced by Streptomyces; producing organisms and improved production. J Antibiot 44:1045–1053
Jones KL (1949) Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J Bacteriol 57:141
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro H (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kaempfer P (2012) Systematics of prokaryotes: the state of the art. Antonie Van Leeuwenhoek 101:3–11
Kelly K (1964) Color-name charts illustrated with centroid colors. Inter-Society Color Council-National Bureau of Standards, Chicago
Kroppenstedt RM, Goodfellow M (2006) The family Thermomonosporaceae: Actinocorallia, Actinomadura, Spirillospora and Thermomonospora. In: Dworkin M, Falkow S, Schleifer KH, Stackebrandt E (eds) The Prokaryotes. Archaea and Bacteria: Firmicutes, Actinomycetes, vol 3, 3rd edn. Springer, New York, pp 682–724
Lai H, Wei X, Jiang Y, Chen X, Li Q, Jiang Y, Jiang C et al (2014) Halopolyspora alba gen. nov., sp. nov., isolated from sediment. Int J Syst Evol Microbiol 64:2775–2780
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Evol Microbiol 20:435–443
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013a) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol R 77:112–143
Liu H, Xiao L, Wei J, Schmitz JC, Liu M, Wang C, Cheng L et al (2013b) Identification of Streptomyces sp. nov. WH26 producing cytotoxic compounds isolated from marine solar saltern in China. World J Microbiol Biotechnol 29:1271–1278
Liu Y, Xie Q-Y, Shi W, Li L, An J-Y, Zhao Y-M, Hong K (2014) Brachybacterium huguangmaarense sp. nov., isolated from Lake sediment. Int J Syst Evol Microbiol 64:1673–1678
Loria R, Kers J, Joshi M (2006) Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol 44:469–487
Lu C, Shen Y (2004) Two new macrolides produced by Streptomyces sp CS. J Antibiot 57(9):597–600
Maciejewska M, Pessi IS, Arguelles-Arias A, Noirfalise P, Luis G, Ongena M, Barton H et al (2015) Streptomyces lunaelactis sp. nov., a novel ferroverdin A-producing Streptomyces species isolated from a moonmilk speleothem. Antonie Van Leeuwenhoek 107:519–531
Malviya N, Yandigeri MS, Yadav AK, Solanki MK, Arora DK (2014) Isolation and characterization of novel alkali-halophilic actinomycetes from the Chilika brackish water lake, India. Ann Microbiol 64:1829–1838
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013a) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC bioinform 14:1
Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P (2013b) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418
Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, Rohde C et al (2014) Complete genome sequence of DSM 30083 T, the type strain (U5/41 T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci 9:1
Minnikin D, O’donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Parte AC (2014) LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616
Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A (2010) How many bootstrap replicates are necessary? J Comput Biol 17:337–354
Quintana ET, Wierzbicka K, Mackiewicz P, Osman A, Fahal AH, Hamid ME, Zakrzewska-Czerwinska J et al (2008) Streptomyces sudanensis sp. nov., a new pathogen isolated from patients with actinomycetoma. Antonie Van Leeuwenhoek 93:305–313
Ray L, Mishra SR, Panda AN, Rastogi G, Pattanaik AK, Adhya TK, Suar M et al (2014) Streptomyces barkulensis sp. nov., isolated from an estuarine lake. Int J Syst Evol Microbiol 64:1365–1372
Russell NJ (1989) Adaptive modifications in membranes of halotolerant and halophilic microorganisms. J Bioenerg Biomembr 21:93–113
Sahay H, Mahfooz S, Singh AK, Singh S, Kaushik R, Saxena AK, Arora DK (2012) Exploration and characterization of agriculturally and industrially important haloalkaliphilic bacteria from environmental samples of hyper saline Sambhar lake, India. World J Microbiol Biotechnol 28:3207–3217
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Santhanam R, Okoro CK, Rong X, Huang Y, Bull AT, Weon H-Y, Andrews BA et al (2012) Streptomyces atacamensis sp. nov., isolated from an extreme hyper-arid soil of the Atacama Desert, Chile. Int J Syst Evol Microbiol 62:2680–2684
Sasser M (1990) Identification of bacteria through fatty acid analysis. In: Klement Z, Rudolph K, Sands D (eds) Methods in phytobacteriology. Akademiai Kiado, Budapest, pp 199–204
Sharma TK, Mawlankar R, Sonalkar VV, Shinde VK, Zhan J, Li W-J, Rele MV et al (2016) Streptomyces lonarensis sp. nov., isolated from Lonar Lake, a meteorite salt water lake in India. Antonie Van Leeuwenhoek 109:225–235
Sharmin T, Rahman MA, Anisuzzaman ASM, Islam MA-U (2013) Antimicrobial and cytotoxic activities of secondary metabolites obtained from a novel species of Streptomyces. Bangladesh Pharm J 16:15–19
Sierra G (1957) A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 23:15–22
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Swofford D (2002) PAUP* version 4.0 b10. Phylogenetic analysis using parsimony (* and other methods). Sinauer, Sunderland, MA
Také A, Matsumoto A, Ōmura S, Takahashi Y (2015) Streptomyces lactacystinicus sp. nov. and Streptomyces cyslabdanicus sp. nov., producing lactacystin and cyslabdan, respectively. J Antibiot 68:322–327
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tatar D, Guven K, Sproer C, Klenk H-P, Sahin N (2014) Streptomyces iconiensis sp. nov. and Streptomyces smyrnaeus sp. nov., two halotolerant actinomycetes isolated from a salt lake and saltern. Int J Syst Evol Microbiol 64:3126–3133
Veyisoglu A, Sahin N (2014) Streptomyces hoynatensis sp. nov., isolated from deep marine sediment. Int J Syst Evol Microbiol 64:819–826
Veyisoglu A, Sahin N (2015) Streptomyces klenkii sp. nov., isolated from deep marine sediment. Antonie Van Leeuwenhoek 107:273–279
Waksman S, Henrici A (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46:337–341
Wayne L, Brenner D, Colwell R, Grimont P, Kandler O, Krichevsky M, Moore L et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Williams S, Goodfellow M, Alderson G, Wellington E, Sneath P, Sackin M (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Xia Z-F, Ruan J-S, Huang Y, Zhang L-L (2013) Streptomyces aidingensis sp. nov., an actinomycete isolated from lake sediment. Int J Syst Evol Microbiol 63:3204–3208
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang X, Liu C, Zhang Y, Wang H, Wang S, Li C, Wang X et al (2014) Streptosporangium shengliensis sp. nov., a novel actinomycete isolated from a lake sediment. Antonie Van Leeuwenhoek 105:237–243
Acknowledgements
HA was supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK). IN was supported by Newcastle University postdoctoral fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2017_970_MOESM1_ESM.docx
Fig. S1 Polar lipid profile of strain MKSP12T. Key: DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; AL, unidentified amino-lipid; GPL, unidentified glycophospholipid; PL1-2, unidentified phospholipids; GL 1-8, unidentified glycolipids. Supplementary material 1 (DOCX 911 kb)
10482_2017_970_MOESM2_ESM.docx
Fig. S2 Neighbour joining phylogenetic tree based on 16S rRNA gene sequences showing relationships between strain MKSP12T and the type strains of closely related Streptomyces species. Actinomadura rifamycini IFO 14183T was used as outgroup. The numbers above the branches are support values when larger than 50% from bootstrapping. The branches are scaled in terms of the expected number of substitutions per site. Supplementary material 2 (DOCX 150 kb)
10482_2017_970_MOESM3_ESM.docx
Table S1 Cellular fatty acid composition (%) of strain MKSP12T and closely related Streptomyces species grown on ISP 2 agar for 10 days at 28 °C. Supplementary material 3 (DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Ay, H., Nouioui, I., del Carmen Montero-Calasanz, M. et al. Streptomyces sediminis sp. nov. isolated from crater lake sediment. Antonie van Leeuwenhoek 111, 493–500 (2018). https://doi.org/10.1007/s10482-017-0970-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0970-z