Abstract
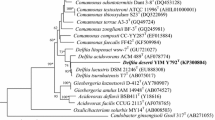
A Gram-negative, strictly aerobic, yellow-pigmented and rod-shaped bacterium, designated strain NSL10T, was isolated from the waste surface soil of a chemical factory in Hongan, China. Strain NSL10T was found to grow optimally at pH 7.0, 30 °C and in the absence of NaCl in modified LB medium. Cells were found to be positive for catalase and oxidase. The G+C content of the total DNA was determined to be 66.8 mol%. The 16S rRNA gene sequence of strain NSL10T showed the highest similarity to that of Devosia albogilva IPL15T (96.80 %), followed by Devosia geojensis BD-c194T (96.46 %) and Devosia chinhatensis IPL18T (96.27 %). The major cellular fatty acids of strain NSL10T were identified as C18:1 ω7c/C18:1 ω6c (48.2 %) and C16:0 (17.7 %). The major polar lipids were identified as diphosphatidylglycerol, phosphatidylglycerol, two unidentified glycolipids and an unidentified compound. Minor amounts of unidentified glycolipids and unidentified polar lipids were also detected. These chemotaxonomic data supported the affiliation of strain NSL10T to the genus Devosia. In conclusion, on the basis of biochemical, physiological characteristics and molecular properties, strain NSL10T represents a novel species within the genus Devosia, for which the name Devosia honganensis sp. nov., is proposed. The type strain is NSL10T (=KCTC 42281T = ACCC 19737T).

Similar content being viewed by others
References
Bertani G (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300
Beveridge TJ, Lawrence JR, Murray RGE (2007) Sampling and staining for light microscopy. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology, 3rd edn. American Society of Microbiology, Washington, DC, pp 19–33
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol 48:459–470
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Foster JW (1944) Microbiological aspects of riboflavina. I. Introduction. II. Bacterial oxidation of riboflavina to lumichrome. J Bacteriol 47:27–41
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH (2012) Introducing EzTaxon-e:a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kluge AG, Farris FS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361
Lane DL (1991) 16S/23S rRNA sequencing. In: Stackebrandt ER, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Liu XM, Chen K, Meng C, Zhang L, Zhu JC, Huang X, Li SP, Jiang JD (2014) Pseudoxanthobacter liyangensis sp. nov., isolated from dichlorodiphenyltrichloroethane-contaminated soil. Int J Syst Evol Microbiol 64:3390–3394
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymock D, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyacids. J Clin Microbiol 16:584–586
Nakagawa Y, Sakane T, Yokota A (1996) Transfer of ‘‘Pseudomonas riboflavina’’ (Foster 1944), a gram-negative, motile rod with long-chain 3-hydroxy fatty acids, to Devosia riboflavina gen. nov., sp. nov., nom. rev. Int J Syst Bacteriol 46:16–22
Parte AC (2013) LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 2013:1–4
Rivas R, Willems A, Subba-Rao NS, Mateos PF, Dazzo FB, Kroppenstedt RM, Martínez-Molina E, Gillis M, Velázquez E (2003) Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst Appl Microbiol 26:47–53
Ryu SH, Chung BS, Le NT, Jang HH, Yun P-Y, Park W, Jeon CO (2008) Devosia geojensis sp. nov., isolated from dieselcontaminated soil in Korea. Int J Syst Evol Microbiol 58:633–636
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using Maximum Likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ, Sikorski J, Smibert RM, Kreig NR (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Marzluf JA, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology, 3rd edn. American Society of Microbiology, Washington, DC, pp 330–393
Vanparys B, Heylen K, Lebbe L, De Vos P (2005) Devosia limi sp. nov., isolated from a nitrifying inoculum. Int J Syst Evol Microbiol 55:1997–2000
Verma M, Kumar M, Dadhwal M, Kaur J, Lal R (2009) Devosia albogilva sp. nov. and Devosia crocina sp. nov., isolated from a hexachlorocyclohexane dump site. Int J Syst Evol Microbiol 59:795–799
Zhang J, Gu T, Zhou Y, He J, Zheng LQ, Li WJ, Huang X, Li SP (2012) Terrimonas rubra sp. nov., isolated from a polluted farmland soil and emended description of the genus Terrimonas. Int J Syst Evol Microbiol 62:2593–2597
Acknowledgments
This work was supported by the Scientific Foundation from Yunnan Tobacco Company (2014YN08), the National Natural Science Foundation of China (J1210056 and J1310015), the Outstanding Youth Foundation of Jiangsu Province (BK20130029) and the Fundamental Research Funds for the Central Universities (KYZ201422).
Author information
Authors and Affiliations
Corresponding authors
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain NSL10T is KP339871.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Song, M., Chen, XL. et al. Devosia honganensis sp. nov., isolated from the soil of a chemical factory. Antonie van Leeuwenhoek 108, 1301–1307 (2015). https://doi.org/10.1007/s10482-015-0582-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0582-4