Abstract
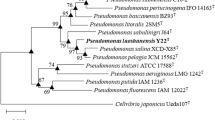
The strain NEAU-ST5-5T was isolated from the saline and alkaline soil in Songnen Plain, North East of China. The bacterium was found to be aerobic, Gram-stain negative, rod-shaped and motile by means of several polar flagella. It forms yellow-orange colonies with a radial wrinkled surface. Phylogenetic analyses based on the separate 16S rRNA gene sequences and concatenated 16S rRNA, gyrB and rpoD gene sequences indicated that it belongs to the genus Pseudomonas in the class Gammaproteobacteria. Strain NEAU-ST5-5T shows gene sequence similarities of 98.8–97.1 % for 16S rRNA, 90.5–78.4 % for gyrB and 90.4–71.1 % for rpoD with type strains of the closely related species of the genus Pseudomonas, respectively. DNA–DNA hybridization relatedness between strain NEAU-ST5-5T and type strains of the most closely related species, Pseudomonas stutzeri DSM 5190T, P. xanthomarina DSM 18231T, P. kunmingensis CGMCC 1.12273T, P. alcaliphila DSM 17744T and P. oleovorans subsp. lubricantis DSM 21016T were 43 ± 1 to 25 ± 2 %. The major fatty acids (>10 %) were determined to be C18:1 ω7c/C18:1 ω6c, C16:1 ω7c/C16:1 ω6c and C16:0, the predominant respiratory quinone was identified as ubiquinone 9 and polar lipids were found to consist of phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, one unknown phospholipid, one unidentified aminophospholipid and one unknown lipid. The genotypic, chemotaxonomic and phenotypic analysis indicated that strain NEAU-ST5-5T represents a novel species of the genus Pseudomonas, for which the name Pseudomonas songnenensis sp. nov. is proposed. The type strain is NEAU-ST5-5T (=ACCC 06361T = DSM 27560T).


Similar content being viewed by others
References
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50:1563–1589. doi:10.1099/00207713-50-4-1563
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142. doi:10.1111/j.1432-1033.1970.tb00830.x
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.2307/2408678
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192. doi:10.1016/S0723-2020(83)80048-4
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi:10.1099/ijs.0.038075-0
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
López JR, Diéguez AL, Doce A, De la Roca E, De la Herran R, Navas JI, Toranzo AE, Romalde JL (2012) Pseudomonas baetica sp. nov., a novel fish pathogen isolated from wedge sole, Dicologoglossa cuneata (Moreau). Int J Syst Evol Microbiol 62:874–882. doi:10.1099/ijs.0.030601-0
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218. doi:10.1016/S0022-2836(61)80047-8
Marmur J, Doty P (1962) Determination of the base composition of Deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118. doi:10.1016/S0022-2836(62)80066-7
Migula W (1894) Uber ein neues System der Bakterien. Arb Bakteriol Inst Karlsruhe 1:235–238 (in German)
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. doi:10.1016/0167-7012(84)90018-6
Palleroni NJ (1984) Genus I. Pseudomonas Migula 1894, 237AL (nom. cons. Opin. 5, Jud. Comm. 1952, 237). In: Krieg NR, Holt JG (ed). Bergey’s manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, p 141–199
Palleroni NJ (1992) Introduction to the family Pseudomonadaceae. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd edn. Springer, New York, pp 3071–3085
Palleroni NJ (1993) Pseudomonas classification. A new case history in the taxonomy of gram-negative bacteria. Antonie Van Leeuwenhoek 64:231–251. doi:10.1007/BF00873084
Palleroni NJ (2003) Prokaryote taxonomy of the 20th century and the impact of studies on the genus Pseudomonas: a personal view. Microbiology 149:1–7. doi:10.1099/mic.0.25952-0
Palleroni NJ (2005) Genus I. Pseudomonas Migula 1894, 237AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (ed). Bergey’s manual of systematic bacteriology, 2nd edn, vol. 2, part B. Springer, NewYork, p 323–379
Pan Y, Huang H, Meng J, Xiao H, Li C, Meng L, Hong S, Liu H, Wang X, Jiang J (2012) Biodiversity of the culturable halotolerant and halophilic bacteria isolated from saline–alkaline soils in Songnen Plain. Acta Microbiol Sin 52:1187–1194
Parte AC (2014) LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616. doi:10.1093/nar/gkt1111
Robinson RA, Stokes RH (1968) Electrolyte solutions, the measurement and interpretation of conductance, chemical potential, and diffusion in solutions of simple electrolytes, 2nd edn. Butterworths, London
Romanenko LA, Uchino M, Falsen E, Lysenko AM, Zhukova NV, Mikhailov VV (2005) Pseudomonas xanthomarina sp. nov., a novel bacterium isolated from marine ascidian. J Gen Appl Microbiol 51:65–71. doi:10.2323/jgam.51.65
Saha R, Spröer C, Beck B, Bagley S (2010) Pseudomonas oleovorans subsp. lubricantis subsp. nov., and reclassification of Pseudomonas pseudoalcaligenes ATCC 17440T as later synonym of Pseudomonas oleovorans ATCC 8062T. Curr Microbiol 60:294–300. doi:10.1007/s00284-009-9540-6
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc, Newark
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Timmis KN (2002) Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol 4:779–781. doi:10.1046/j.1462-2920.2002.00365.x
Vancanneyt M, Witt S, Abraham WR, Kersters K, Fredrickson HL (1996) Fatty acid content in whole-cell hydrolysates and phospholipid fractions of Pseudomonas: a taxonomic evaluation. Syst Appl Microbiol 19:528–540. doi:10.1016/S0723-2020(96)80025-7
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464. doi:10.1099/00207713-37-4-463
Xie FH, Ma H, Quan SJ, Liu DH, Chen GC, Chao YP, Qian SJ (2014) Pseudomonas kunmingensis sp. nov., an exopolysaccharide-producing bacterium isolated from a phosphate mine. Int J Syst Evol Microbiol 64:559–564. doi:10.1099/ijs.0.055632-0
Yamamoto S, Harayama S (1995) PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol 61:1104–1109
Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S (2000) Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556. doi:10.1093/bioinformatics/13.5.555
Yumoto I, Yamazaki K, Hishinuma M, Nodasaka Y, Suemori A, Nakajima K, Inoue N, Kawasaki K (2001) Pseudomonas alcaliphila sp. nov., a novel facultatively psychrophilic alkaliphile isolated from seawater. Int J Syst Evol Microbiol 51:349–355. doi:10.1099/00207713-51-2-349
Zhou Y, Dong J, Wang X, Huang X, Zhang KY, Zhang YQ, Guo YF, Lai R, Li WJ (2007) Chryseobacterium flavum sp. nov., isolated from polluted soil. Int J Syst Evol Microbiol 57:1765–1769. doi:10.1099/ijs.0.65046-0
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 31370087) and Natural Science Foundation of Heilongjiang Province of China (Grant No. C201417).
Author information
Authors and Affiliations
Corresponding author
Additional information
Lei Zhang, Yuanyuan Pan and Kaibiao Wang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Pan, Y., Wang, K. et al. Pseudomonas songnenensis sp. nov., isolated from saline and alkaline soils in Songnen Plain, China. Antonie van Leeuwenhoek 107, 711–721 (2015). https://doi.org/10.1007/s10482-014-0365-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0365-3