Abstract
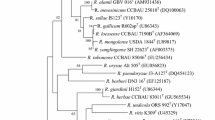
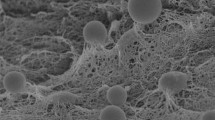
During a study of endophytic bacteria from traditional Chinese medicinal plants, a bacterial strain, designated PTYR-5T, was isolated from the leaf of Smilacina japonica A. Gray collected from Taibai Mountain in Shaanxi Province, north-west China. Phylogenetic analysis based on 16S rRNA gene sequences showed that strain PTYR-5T is a member of the genus Rhizobium, exhibiting the highest sequence similarities to R. cellulosilyticum LMG 23642T (97.2 %), R. huautlense LMG 18254T (97.2 %) and R. alkalisoli CCBAU 01393T (97.1 %). The levels of 16S rRNA gene sequence similarity with respect to other Rhizobium species with validly published names were less than 97.0 %. Phylogenies of the housekeeping genes atpD, recA and glnII confirmed its distinct position, showing low similarity with respect to those of recognized Rhizobium species (no more than 94.1 , 90.0 and 88.0 % similarity, respectively). The DNA–DNA relatedness values of strain PTYR-5T with R. cellulosilyticum LMG 23642T, R. huautlense LMG 18254T and R. alkalisoli CCBAU 01393T were 33.6, 21.4 and 29.5 %, respectively. Based on phenotypic, phylogenetic and genotypic data, strain PTYR-5T is considered to represent a novel species of the genus Rhizobium, for which the name Rhizobium smilacinae sp. nov. is proposed. The type strain is PTYR-5T (=CCTCC AB 2013016T=KCTC 32300T=LMG 27604T).

Similar content being viewed by others
References
Amarger N, Macheret V, Laguerre G (1997) Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int J Syst Bacteriol 47:996–1006
Bibi F, Chung EJ, Khan A, Jeon CO, Chung YR (2012) Rhizobium halophytocola sp. nov., isolated from the root of a coastal dune plant. Int J Syst Evol Microbiol 62:1997–2003
Cleenwerck I, Vandemeulebroecke K, Janssens D, Swings J (2002) Re-examination of the genus Acetobacter, with descriptions of Acetobacter cerevisiae sp. nov. and Acetobacter malorum sp. nov. Int J Syst Evol Microbiol 52:1551–1558
Doetsch RN (1981) Determinative methods of light microscopy. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GH (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 21–33
Euzéby JP (2014) List of prokaryotic names with standing in nomenclature. http://www.bacterio.cict.fr/
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid–deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Frank B (1889) Über die Pilzsymbiose der Leguminosen. Ber Dtsch Bot Ges 7:332–346 (in German)
Fujita H, Aoki S, Kawaguchi M (2014) Evolutionary dynamics of nitrogen fixation in the legume–rhizobia symbiosis. PLoS ONE 9(4):e93670
Gao JL, Sun JG, Li Y, Wang ET, Chen WX (1994) Numerical taxonomy and DNA relatedness of tropical rhizobia isolated from Hainan Province, China. Int J Syst Bacteriol 44:151–158
García-Fraile P, Rivas R, Willems A, Peix A, Martens M, Martínez-Molina E, Mateos PF, Velázquez E (2007) Rhizobium cellulosilyticum sp. nov., isolated from sawdust of Populus alba. Int J Syst Evol Microbiol 57:844–848
Gaunt MW, Turner SL, Rigottier-Gois L, Lloyd-Macgilp SA, Young JPW (2001) Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol 51:2037–2048
Hunter WJ, Kuykendall LD, Manter DK (2007) Rhizobium selenireducens sp. nov. a selenite-reducing α-Proteobacteria isolated from a bioreactor. Curr Microbiol 55:455–460
Kim BC, Poo H, Lee KH, Kim MN, Kwon OY, Shin KS (2012) Mucilaginibacter angelicae sp. nov., isolated from the rhizosphere of Angelica polymorpha Maxim. Int J Syst Evol Microbiol 62:55–60
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kittiwongwattana C, Thawai C (2013) Rhizobium paknamense sp. nov., isolated from lesser duckweeds (Lemna aequinoctialis). Int J Syst Evol Microbiol 63:3823–3828
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981–993
Lane DJ (1991) 16S-23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 125–175
López-López A, Rogel MA, Ormeño-Orrillo E, Martínez-Romero J, Martínez-Romero E (2010) Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst Appl Microbiol 33:322–327
Lu LY, Chen WF, Han LL, Wang ET, Chen WX (2009) Rhizobium alkalisoli sp. nov., isolated from Caragana intermedia growing in saline-alkaline soils in the north of China. Int J Syst Evol Microbiol 59:3006–3011
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Peng G, Yuan Q, Li H, Zhang W, Tan Z (2008) Rhizobium oryzae sp. nov., isolated from the wild rice Oryza alta. Int J Syst Evol Microbiol 58:2158–2163
Quan ZX, Bae HS, Baek JH, Chen WF, Im WT, Lee ST (2005) Rhizobium daejeonense sp. nov., isolated from a cyanide treatment bioreactor. Int J Syst Evol Microbiol 55:2543–2549
Rosenblueth M, Martinez-Romero E (2004) Rhizobium etli maize populations and their competitiveness for root colonization. Arch Microbiol 181:337–344
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc, Newark
Schloter M, Wiehe W, Assmus B, Steindl H, Becke H, Höflich G, Hartmann A (1997) Root colonization of different plants by plant-growth-promoting Rhizobium leguminosarum bv. trifolii R39 studied with monospecific polyclonal antisera. Appl Environ Microbiol 63:2038–2046
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludeña D, Mateos PF, Martínez-Molina E, Velázquez E (2005) Nodulation of Lupinus albus by strains of Ochrobactrum lupine sp. nov. Appl Environ Microbiol 71:1318–1327
Turner SL, Young JPW (2000) The glutamine synthetases of rhizobia: phylogenetics and evolutionary implications. Mol Biol Evol 17:309–319
Vincent JM (1970) The cultivation, isolation and maintenance of rhizobia. In: Vincentl JM (ed) A manual for the practical study of the root-nodule bacteria. Blackwell, Oxford, pp 1–13
Wang ET, van Berkum P, Beyene D, Sui XH, Dorado O, Chen WX, Martinez-Romero E (1998) Rhizobium huautlense sp. nov., a symbiont of Sesbania herbacea that has a close phylogenetic relationship with Rhizobium galegae. Int J Syst Bacteriol 48:687–699
Wang F, Wang ET, Wu LJ, Sui XH, Li Y Jr, Chen WX (2011) Rhizobium vallis sp. nov., isolated from nodules of three leguminous species. Int J Syst Evol Microbiol 61:2582–2588
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wilson K (1987) Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, pp 241–245
Yoon JH, Kang SJ, Yi HS, Oh TK, Ryu CM (2010) Rhizobium soli sp. nov., isolated from soil. Int J Syst Evol Microbiol 60:1387–1393
Young JM, Kuykendall LD, Martínez-Romero E, Kerr A, Sawada H et al (2001) A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie, 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int J Syst Evol Microbiol 51:89–103
Zhang L, Wang Y, Wei L, Wang Y, Shen X, Li S (2013) Taibaiella smilacinae gen. nov., sp. nov., an endophytic member of the family Chitinophagaceae isolated from the stem of Smilacina japonica, and emended description of Flavihumibacter petaseus. Int J Syst Evol Microbiol 63:3769–3776
Zhang L, Wei L, Zhu L, Li C, Wang Y, Shen X (2014) Pseudoxanthomonas gei sp. nov., a novel endophytic bacterium isolated from the stem of Geum aleppicum. Antonie Van Leeuwenhoek 105:653–661
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 program, grant 2013AA102802), the National Natural Science Foundation of China (Grant No. 31100001 and 31170100) and the Natural Science Foundation of Shaanxi Province, China (Grant No. 2012JQ3006).
Author information
Authors and Affiliations
Corresponding author
Additional information
Lei Zhang and Xu Shi have contributed equally to this work.
The GenBank/EMBL/DDBJ accession numbers for the partial 16S rRNA, atpD, recA and glnII gene sequences of strain PTYR-5T are KF551141, KF738707, KF738708 and KF738709, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Shi, X., Si, M. et al. Rhizobium smilacinae sp. nov., an endophytic bacterium isolated from the leaf of Smilacina japonica . Antonie van Leeuwenhoek 106, 715–723 (2014). https://doi.org/10.1007/s10482-014-0241-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0241-1