Abstract
The gut microbiome may modulate intestinal immunity by luminal conversion of dietary amino acids to biologically active signals. The model probiotic organism Lactobacillus reuteri ATCC PTA 6475 is indigenous to the human microbiome, and converts the amino acid l-histidine to the biogenic amine, histamine. Histamine suppresses tumor necrosis factor (TNF) production by human myeloid cells and is a product of l-histidine decarboxylation, which is a proton-facilitated reaction. A transposon mutagenesis strategy was developed based on a single-plasmid nisin-inducible Himar1 transposase/transposon delivery system for L. reuteri. A highly conserved proton-chloride antiporter gene (eriC), a gene widely present in the gut microbiome was discovered by Himar1 transposon (Tn)-mutagenesis presented in this study. Genetic inactivation of eriC by transposon insertion and genetic recombineering resulted in reduced ability of L. reuteri to inhibit TNF production by activated human myeloid cells, diminished histamine production by the bacteria and downregulated expression of histidine decarboxylase cluster genes compared to those of WT 6475. EriC belongs to a large family of ion transporters that includes chloride channels and proton-chloride antiporters and may facilitate the availability of protons for the decarboxylation reaction, resulting in histamine production by L. reuteri. This report leverages the tools of bacterial genetics for probiotic gene discovery. The findings highlight the widely conserved nature of ion transporters in bacteria and how ion transporters are coupled with amino acid decarboxylation and contribute to microbiome-mediated immunomodulation.





Similar content being viewed by others
References
Akerley BJ, Lampe DJ (2002) Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol 358:100–108
Akerley BJ, Rubin EJ, Camilli A, Lampe DJ, Robertson HM, Mekalanos JJ (1998) Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA 95(15):8927–8932
Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR (2004) Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol 70(9):5315–5322. doi:10.1128/AEM.70.9.5315-5322.2004
Beare PA (2012) Genetic manipulation of Coxiella burnetii. Adv Exp Med Biol 984:249–271. doi:10.1007/978-94-007-4315-1_13
Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA (2009) Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 191(5):1369–1381. doi:10.1128/JB.01580-08
Bourhy P, Louvel H, Saint Girons I, Picardeau M (2005) Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J Bacteriol 187(9):3255–3258. doi:10.1128/JB.187.9.3255- 3258.2005
Britton RA, Versalovic J (2008) Probiotics and gastrointestinal infections. Interdiscip Perspect Infect Dis 2008:290769. doi:10.1155/2008/290769
Chen TY (2005) Structure and function of clc channels. Annu Rev Physiol 67:809–839. doi:10.1146/annurev.physiol.67.032003.153012
Cotter PD, Hill C (2003) Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67(3):429–453
De Biase D, Pennacchietti E (2012) Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol. doi:10.1111/mmi.12020
de Vos WM, Kuipers OP, van der Meer JR, Siezen RJ (1995) Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol Microbiol 17(3):427–437
Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian KD (1994) Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol 60(3):814–825
Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NC, Patil PB, Juge N, Mackenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J (2011) The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet 7(2):e1001314. doi:10.1371/journal.pgen.1001314
Gonzaga VE, Lescano AG, Huaman AA, Salmon-Mulanovich G, Blazes DL (2009) Histamine levels in fish from markets in Lima. Peru J Food Prot 72(5):1112–1115
Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6(3):279–289. doi:10.1016/j.chom.2009.08.003
Guillet R, Stoll BJ, Cotten CM, Gantz M, McDonald S, Poole WK, Phelps DL (2006) Association of H2-blocker therapy and higher incidence of necrotizing enterocolitis in very low birth weight infants. Pediatrics 117(2):e137–e142. doi:10.1542/peds.2005-1543
Gury J, Barthelmebs L, Cavin JF (2004) Random transposon mutagenesis of Lactobacillus plantarum by using the pGh9:IS S1 vector to clone genes involved in the regulation of phenolic acid metabolism. Arch Microbiol 182(5):337–345. doi:10.1007/s00203-004-0705-1
Gut H, Pennacchietti E, John RA, Bossa F, Capitani G, De Biase D, Grutter MG (2006) Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. EMBO J 25(11):2643–2651. doi:10.1038/sj.emboj.7601107
Hemarajata P, Gao C, Pflughoeft KJ, Thomas CM, Saulnier DM, Spinler JK, Versalovic J (2013) Lactobacillus reuteri-Specific Immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri. J Bacteriol 195(24):5567–5576. doi:10.1128/JB.00261-13
Howery AE, Elvington S, Abraham SJ, Choi KH, Dworschak-Simpson S, Phillips S, Ryan CM, Sanford RL, Almqvist J, Tran K, Chew TA, Zachariae U, Andersen OS, Whitelegge J, Matulef K, Du Bois J, Maduke MC (2012) A designed inhibitor of a CLC antiporter blocks function through a unique binding mode. Chem Biol 19(11):1460–1470. doi:10.1016/j.chembiol.2012.09.017
Hungerford JM (2010) Scombroid poisoning: a review. Toxicon 56(2):231–243. doi:10.1016/j.toxicon.2010.02.006
Ito M, Kim YG, Tsuji H, Kiwaki M, Nomoto K, Tanaka R, Okada N, Danbara H (2010) A practical random mutagenesis system for probiotic Lactobacillus casei using Tn5 transposition complexes. J Appl Microbiol 109(2):657–666. doi:10.1111/j.1365-2672.2010.04690.x
Iyer R, Iverson TM, Accardi A, Miller C (2002) A biological role for prokaryotic ClC chloride channels. Nature 419(6908):715–718. doi:10.1038/nature01000
Iyer R, Williams C, Miller C (2003) Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol 185(22):6556–6561
Juillerat P, Schneeweiss S, Cook EF, Ananthakrishnan AN, Mogun H, Korzenik JR (2012) Drugs that inhibit gastric acid secretion may alter the course of inflammatory bowel disease. Aliment Pharmacol Ther 36(3):239–247. doi:10.1111/j.1365-2036.2012.05173.x
Kieseritzky G, Knapp EW (2010) Charge transport in the ClC-type chloride-proton anti-porter from Escherichia coli. J Biol Chem 286(4):2976–2986. doi:10.1074/jbc.M110.163246
Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP (1997) Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol 63(11):4581–4584
Kristich CJ, Nguyen VT, Le T, Barnes AM, Grindle S, Dunny GM (2008) Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol 74(11):3377–3386. doi:10.1128/AEM.02665-07
Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270(45):27299–27304
Lampe DJ, Grant TE, Robertson HM (1998) Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149(1):179–187
Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM (1999) Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA 96(20):11428–11433
Landete JM, Pardo I, Ferrer S (2006) Histamine, histidine, and growth-phase mediated regulation of the histidine decarboxylase gene in lactic acid bacteria isolated from wine. FEMS Microbiol Lett 260(1):84–90. doi:10.1111/j.1574-6968.2006.00294.x
Landete JM, Pardo I, Ferrer S (2008) Regulation of hdc expression and HDC activity by enological factors in lactic acid bacteria. J Appl Microbiol 105(5):1544–1551. doi:10.1111/j.1365-2672.2008.03865.x
Larionov A, Krause A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6:62. doi:10.1186/1471-2105-6-62
Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K (1995) A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177(24):7011–7018
Le Breton Y, Mohapatra NP, Haldenwang WG (2006) In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl Environ Microbiol 72(1):327–333. doi:10.1128/AEM.72.1.327-333.2006
Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J (1996) A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253(1–2):217–224
Licandro-Seraut H, Brinster S, van de Guchte M, Scornec H, Maguin E, Sansonetti P, Cavin JF, Serror P (2012) Development of an efficient in vivo system (Pjunc-TpaseIS1223) for random transposon mutagenesis of Lactobacillus casei. Appl Environ Microbiol 78(15):5417–5423. doi:10.1128/AEM.00531-12
Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177(14):4097–4104
Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J (2008) Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis 14(8):1068–1083. doi:10.1002/ibd.20448
Liu H, Bouillaut L, Sonenshein AL, Melville SB (2013) Use of a mariner-based transposon mutagenesis system to isolate Clostridium perfringens mutants deficient in gliding motility. J Bacteriol 195(3):629–636. doi:10.1128/JB.01288-12
Maduke M, Miller C, Mindell JA (2000) A decade of CLC chloride channels: structure, mechanism, and many unsettled questions. Annu Rev Biophys Biomol Struct 29:411–438. doi:10.1146/annurev.biophys.29.1.411
Maier TM, Pechous R, Casey M, Zahrt TC, Frank DW (2006) In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl Environ Microbiol 72(3):1878–1885. doi:10.1128/AEM.72.3.1878-1885.2006
Metz M, Doyle E, Bindslev-Jensen C, Watanabe T, Zuberbier T, Maurer M (2011) Effects of antihistamines on innate immune responses to severe bacterial infection in mice. Int Arch Allergy Immunol 155(4):355–360. doi:10.1159/000321614
Molenaar D, Abee T, Konings WN (1991) Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim Biophys Acta 1115(1):75–83
Naila A, Flint S, Fletcher G, Bremer P, Meerdink G (2011) Control of biogenic amines in food–existing and emerging approaches. J Food Sci 75(7):R139–R150. doi:10.1111/j.1750-3841.2010.01774.x
Nolan RA (1971) Amino acids and growth factors in vitamin-free casamino acids. Mycologia 63(6):1231–1234
Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J (2010) Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J 4(3):377–387. doi:10.1038/ismej.2009.123
O’Mahony L, Akdis M, Akdis CA (2011) Regulation of the immune response and inflammation by histamine and histamine receptors. J Allergy Clin Immunol 128(6):1153–1162. doi:10.1016/j.jaci.2011.06.051
Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, Mercenier A (2000) Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl Environ Microbiol 66(10):4427–4432
Pessione E, Mazzoli R, Giuffrida MG, Lamberti C, Garcia-Moruno E, Barello C, Conti A, Giunta C (2005) A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics 5(3):687–698. doi:10.1002/pmic.200401116
Picardeau M (2009) Transposition of fly mariner elements into bacteria as a genetic tool for mutagenesis. Genetica. doi:10.1007/s10709-009-9408-5
Pozsgai ER, Blair KM, Kearns DB (2011) Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Appl Environ Microbiol 78(3):778–785. doi:10.1128/AEM.07098-11
Que YA, Haefliger JA, Francioli P, Moreillon P (2000) Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect Immun 68(6):3516–3522
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26(1):51–56. doi:10.1016/S0097-8485(01)00099-7
Richard H, Foster JW (2004) Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol 186(18):6032–6041. doi:10.1128/JB.186.18.6032- 6041.2004
Rodwell AW (1953) The histidine decarboxylase of a species of Lactobacillus; apparent dispensability of pyridoxal phosphate as coenzyme. J Gen Microbiol 8(2):233–237
Romano A, Trip H, Lonvaud-Funel A, Lolkema JS, Lucas PM (2012) Evidence of two functionally distinct ornithine decarboxylation systems in lactic acid bacteria. Appl Environ Microbiol 78(6):1953–1961. doi:10.1128/AEM.07161-11
Rosander A, Connolly E, Roos S (2008) Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl Environ Microbiol 74(19):6032–6040. doi:10.1128/AEM.00991-08
Rossi F, Gardini F, Rizzotti L, La Gioia F, Tabanelli G, Torriani S (2011) Quantitative analysis of histidine decarboxylase gene (hdcA) transcription and histamine production by Streptococcus thermophilus PRI60 under conditions relevant to cheese making. Appl Environ Microbiol 77(8):2817–2822. doi:10.1128/AEM.02531-10
Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J (1998) A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27(2):299–310
Sip M, Herman P, Plasek J, Hrouda V (1990) Transmembrane potential measurement with carbocyanine dye diS-C3-(5): fast fluorescence decay studies. J Photochem Photobiol B 4(3):321–328
Su MS, Schlicht S, Ganzle MG (2011) Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb Cell Fact 10 Suppl 1:S8. doi:10.1186/1475-2859-10-S1-S8
Tam C, Glass EM, Anderson DM, Missiakas D (2006) Transposon mutagenesis of Bacillus anthracis strain Sterne using Bursa aurealis. Plasmid 56(1):74–77. doi:10.1016/j.plasmid.2006.01.002
The Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207–214
Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J (2012) Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 7(2):e31951. doi:10.1371/journal.pone.0031951
van Pijkeren JP, Britton RA (2012) High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40(10):e76. doi:10.1093/nar/gks147
van Pijkeren JP, Neoh KM, Sirias D, Findley AS, Britton RA (2012) Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 3(4):209–217. doi:10.4161/bioe.21049
Walter J, Chagnaud P, Tannock GW, Loach DM, Dal Bello F, Jenkinson HF, Hammes WP, Hertel C (2005) A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl Environ Microbiol 71(2):979–986. doi:10.1128/AEM.71.2.979-986.2005
Walter J, Britton RA, Roos S (2010) Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci USA 108 Suppl 1:4645–4652. doi:10.1073/pnas.1000099107
Wells CL, Moore EA, Hoag JA, Hirt H, Dunny GM, Erlandsen SL (2000) Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect Immun 68(12):7190–7194
Wu CM, Lin CF, Chang YC, Chung TC (2006) Construction and characterization of nisin-controlled expression vectors for use in Lactobacillus reuteri. Biosci Biotechnol Biochem 70(4):757–767. doi:http://www.ncbi.nlm.nih.gov/pubmed/16636439
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. doi:10.1101/gr.074492.107
Acknowledgments
This work was supported in part by research support from National Institutes of Health (R01 AT004326, R01 DK065075, UH3 DK083990). We also acknowledge NIH support (DK56338) for the Texas Medical Center Digestive Diseases Center. We thank Eamonn Connolly (BioGaia AB, Stockholm) for providing the L. reuteri strains, David Lampe (Duquesne University, Pittsburgh, PA) and Todd Klaenhammer (North Carolina State University, Raleigh, NC) for providing plasmids used in this study, Kathryn Pflughoeft for designing recombineering oligonucleotides and scientific expertise, and Michael White for his assistance in generating Himar1 Tn-mutants.
Conflict of interest
J Versalovic receives unrestricted research support from Biogaia AB (Stockholm, Sweden).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. S1
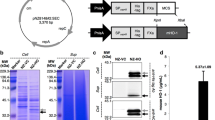
Schematic of single-plasmid Himar1 Transposon mutagenesis in L. reuteri. L. reuteri 6475 was electroporated with pPH-M1 (2 mm cuvettes at 2,500 V, 25 μF and 400 Ω) and transformants were cultured in MRS with Cm 10 and Erm 10 in the presence of nisin overnight at 30 °C to induce for Himar1-transposase expression and transposition. Consecutive subculturing at 45 °C in the absence of Cm and presence of Erm was carried out to cure Himar1 Tn-insertion mutants of pPH-M1. Finally, L. reuteri::Himar1 Tn-mutants were selected for growth on Erm 10 and corresponding susceptibility to Cm 10. L. reuteri::Himar1 Tn-mutants meeting this susceptibility pattern were included in our Tn-mutant library and stored in 96-well plates at −80 °C. (TIFF 13538 kb)
Supplemental Fig. S2
Southern hybridization demonstrated the presence of Himar1 Tn in L. reuteri chromosomal DNA. HindIII digestions of gDNA from 16 randomly chosen Tn-mutants (lanes 1–16) were subjected to Southern hybridization with a DIG-labeled probe specific to ermR in the Himar1 Tn. Bands in each lane represent the presence of the Himar1 Tn in DNA fragments. DIG-labeled molecular weight marker VII (Roche, Indianapolis, IN) is shown on the left and are indicated in bp. “+” positive control/HindIII-digested pPH-M1, “−” negative control/HindIII-digested WT 6475 gDNA. (TIFF 6688 kb)
Supplemental Fig. S3
Preliminary high-throughput TNF inhibitory screening of L. reuteri::Himar1 Tn-mutants. Conditioned media from Himar1 Tn-mutants were tested for the ability to inhibit TNF by TLR2-activated THP-1 cells in a 96-well format. L.reuteri::Himar1 Tn-mutants with ≥50 % reduction in TNF suppression as compared to wild type 6475 were selected for phenotype confirmation in the full-scale TNF inhibition assay. The results are expressed as the mean ± SD, n=3, *p value < 0.01, **p value < 0.005 compared to WT 6475 using unpaired t test. Data from 72 of the 216 screened L. reuteri::Himar1 mutants are shown here. Data from the remaining 144 L. reuteri::Himar1 Tn mutants are not shown. (TIFF 17377 kb)
Supplemental Fig. S4
A proposed model of how EriC regulates histidine decarboxylation in L. reuteri 6475. We propose that EriC simultaneously coordinates the import of protons with the export of electrons to maintain a neutral membrane potential, and is constantly relieving the inside-negative membrane potential that accumulates during histidine decarboxylation. This process would also replenish intracellular proton concentrations required to continue producing histamine. HdcB a putative helper protein proposed to convert HdcA into a mature enzyme, is not depicted. HdcA histidine decarboxylase enzyme, HdcP histidine-histamine antiporter. (TIFF 7005 kb)
Rights and permissions
About this article
Cite this article
Hemarajata, P., Spinler, J.K., Balderas, M.A. et al. Identification of a proton-chloride antiporter (EriC) by Himar1 transposon mutagenesis in Lactobacillus reuteri and its role in histamine production. Antonie van Leeuwenhoek 105, 579–592 (2014). https://doi.org/10.1007/s10482-014-0113-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0113-8