Abstract
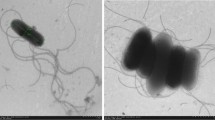
A Gram stain-negative, aerobic and rod-shaped bacterium, strain DY22T, was isolated from a deep-sea sediment collected from the east Pacific Ocean. The isolate was found to grow in the presence of 0–20.0 % (w/v) NaCl and at pH 4.5–8.5; optimum growth was observed with 0.5–2.0 % (w/v) NaCl and at pH 5.0–7.0. Chemotaxonomic analysis showed the presence of ubiquinone-9 as predominant respiratory quinone and C16:0, C19:0 ω8c cyclo and C12:0 3-OH as major cellular fatty acids. The genomic DNA G+C content was determined to be 59.6 mol%. Comparative 16S rRNA gene sequence analysis revealed that the novel isolate belongs to the genus Salinicola. Strain DY22T exhibited the closest phylogenetic affinity to the type strain of Salinicola salarius with 97.2 % sequence similarity and less than 97 % sequence similarity with respect to other Salinicola species with validly published names. The DNA–DNA reassociation values between strain DY22T and S. salarius DSM 18044T was 52 ± 4 %. On the basis of phenotypic, chemotaxonomic and genotypic data, strain DY22T represents a novel species of the genus Salinicola, for which the name Salinicola peritrichatus sp. nov. (type strain DY22T = CGMCC 1.12381T = JCM 18795T) is proposed.


Similar content being viewed by others
References
Aguilera M, Cabrera A, Incerti C et al (2007) Chromohalobacter salarius sp. nov., a moderately halophilic bacterium isolated from a solar saltern in Cabo de Gata, Almería, southern Spain. Int J Syst Evol Microbiol 57:1238–1242
Anańina LN, Plotnikova EG, Gavrish E et al (2007) Salinicola socius gen. nov., sp. nov., a moderately halophilic bacterium from a naphthalene-utilizing microbial association. Mikrobiologiia 76:369–376 Microbiology (Engl. Transl.) 76:324–330
Arahal DR, Vreeland RH, Litchfield CD et al (2007) Recommended minimal standards for describing new taxa of the family Halomonadaceae. Int J Syst Evol Microbiol 57:2436–2446
de la Haba RR, Sánchez-Porro C, Márquez MC et al (2010) Taxonomic study of the genus Salinicola: transfer of Halomonas salaria and Chromohalobacter salarius to the genus Salinicola as Salinicola salarius comb. nov. and Salinicola halophilus nom. nov., respectively. Int J Syst Evol Microbiol 60:963–971
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Huß VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Kim KK, Jin L, Yang HC et al (2007) Halomonas gomseomensis sp. nov., Halomonas janggokensis sp. nov., Halomonas salaria sp. nov. and Halomonas denitrificans sp. nov., moderately halophilic bacteria isolated from saline water. Int J Syst Evol Microbiol 57:675–681
Kim O-S, Cho Y-J, Lee K et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Leifson E (1963) Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85:1183–1184
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Mata JA, Martínez-Cánovas J, Quesada E et al (2002) A detailed phenotypic characterisation of the type strains of Halomonas species. Syst Appl Microbiol 25:360–375
Mesbah M, Whitman WB (1989) Measurement of deoxyguanosine/thymidine ratios in complex mixtures by high-performance liquid chromatography for determination of the mole percentage guanine + cytosine of DNA. J Chromatogr 479:297–306
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22:4673–4680
Ventosa A, Quesada E, Rodriguez-Valera F et al (1982) Numerical taxonomy of moderately halophilic Gram-negative rods. J Gen Microbiol 128:1959–1968
Wayne LG, Brenner DJ, Colwell RR et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Xu X-W, Wu Y-H, Zhou Z et al (2007) Halomonas saccharevitans sp. nov., Halomonas arcis sp. nov. and Halomonas subterranea sp. nov., halophilic bacteria isolated from hypersaline environments of China. Int J Syst Evol Microbiol 57:1619–1624
Xu X-W, Huo Y–Y, Bai X-D et al (2011) Kordiimonas lacus sp. nov., isolated from a ballast water tank, and emended description of the genus Kordiimonas. Int J Syst Evol Microbiol 61:422–426
Acknowledgments
We thank Jean Euzéby for his help with the specific etymology and nomenclature. This study was supported by Grants from China Ocean Mineral Resources R & D Association (COMRA) Special Foundation (DY125-14-E-02), the Public Science and Technology Research Funds Projects of Ocean (201005032-3) and the National Natural Science Foundation of China (41276173).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank accession number for 16S rRNA gene sequence of S. peritrichatus DY22T is KC005304.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huo, YY., Meng, FX., Xu, L. et al. Salinicola peritrichatus sp. nov., isolated from deep-sea sediment. Antonie van Leeuwenhoek 104, 55–62 (2013). https://doi.org/10.1007/s10482-013-9925-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9925-1