Abstract
Livestock diseases such as bovine tuberculosis can have considerable negative effects on human health and economic activity. Wildlife reservoirs often hinder disease eradication in sympatric livestock populations. Therefore, quantifying interactions between wildlife and livestock is an important aspect of understanding disease persistence. This study was conducted on an extensive cattle farm in southwest Spain, where cattle, domestic pigs, wild boar and red deer are considered to be part of a tuberculosis host community. We tested the hypothesis that the frequency of both types of interactions would be greater at food and water sites, due to the aggregation of individuals from multiple species at these locations. We measured direct and indirect interactions between individuals using GPS and proximity loggers. Over 57,000 direct interactions were recorded over a 2-year period, of which 875 (1.5 %) occurred between different species and 216 (0.38 %) occurred between wildlife and livestock. Most direct and indirect interactions occurred at water sites. Over 90 % of indirect interactions between wildlife and livestock took place within the estimated 3-day environmental survival time of Mycobacterium bovis in this habitat. Red deer home ranges and daily activity patterns revealed significant spatial and temporal overlaps with cattle, particularly in autumn. Suids and red deer also cross the farm boundary regularly, introducing a between-farm interaction risk. The infrequent occurrence of direct interactions between individuals from different species suggests that they are unlikely to be the sole mode of disease transmission and that indirect interactions may play an important role.
Similar content being viewed by others
Introduction
Livestock diseases pose a significant threat to human health, social wellbeing and economic activity (Michel et al. 2010; Schiller et al. 2011). Over 77 % of the pathogens affecting domestic mammals can infect multiple hosts (Cleaveland et al. 2001; Martin et al. 2011). This can result in epidemiologically connected multi-species communities in which a pathogen persists (Haydon et al. 2002). Disease transmission depends on the characteristics and virulence of the pathogen and how long it can survive in the environment, as well as the ecology, susceptibility and pathogen excretion rates of each host species, existing disease prevalence, environmental conditions and contact between infected and susceptible animals (Anderson and May 1979; Martin et al. 2011; Craft et al. 2011; Vander Wal et al. 2012). Social behaviour strongly influences interactions between individuals of the same and different species (Böhm et al. 2008; Drewe 2010). This study focusses on spatially and temporally quantifying the contacts between individuals from multiple species in a disease community. Identifying the different roles each species plays in the transmission of a disease will improve the evidence base for making decisions about strategies for effective disease management.
Inter-species transmission of disease may occur through direct or indirect interactions between individuals. Direct interaction refers to direct physical contact or very close proximity between individuals. In a direct interaction, a pathogen may be passed between individuals with little or no exposure to the environment, e.g. physical contact or inhalation of aerosol particles from the exhalation of an infected individual. Indirect interaction refers to shared space use, where individuals visit the same location at different times. They are likely to be driven by intake behaviour such as feeding or drinking (Hutchings and Harris 1997), particularly if food and water resources are not evenly distributed, causing aggregation of multiple species (Barasona et al. 2014a). Transmission through indirect interaction requires the pathogen to survive exposure to the environment for sufficient time remain infective when encountered by different potential host individuals. Indirect interactions may be important in the transmission of diseases that demonstrate environmental survival (Hutchings and Harris 1999; Drewe et al. 2013) and should be considered in the management of these diseases (Kukeilka et al. 2013).
Proximity data loggers and GPS technology allow for the quantification of direct and indirect interactions (Prange et al. 2006; Walrath 2011; Drewe et al. 2012). To date, only two studies have used proximity loggers to quantify close direct interactions between multiple species; both studying contact between badgers (Meles meles) and cattle (Bos primigenius) in the UK with reference to tuberculosis (TB) transmission. Böhm et al. (2009) identified considerable individual heterogeneity in contact rates and found that direct contacts between badgers and cattle did occur, although they were infrequent. Drewe et al. (2013) found that indirect interactions between species were much more frequent overall than direct interactions. GPS loggers have been used to identify the probability of interaction between cattle and wild boar in Spain (Barasona et al. 2014a), wild and domestic bovids in Africa (Miguel et al. 2013) and between cattle and pigs in Texas (Cooper et al. 2010). They have also shown that proximity between elk (Cervus elaphus), mule deer (Odecoileus hemionus) and cattle in the USA varied with different combinations of species (zu Dohna et al. 2014).
TB is one of the most widespread examples of a disease that is prevalent in both wildlife and livestock (Fitzgerald and Kaneene 2013). Primarily caused in cattle by Mycobacterium bovis infection, this bacterial disease can spill over into human and wildlife populations. Wildlife reservoirs have been identified in brush-tailed possums (Trichosurus vulpecula) in New Zealand (Anderson et al. 2013); badgers in the England, Wales and Ireland (O’Connor et al. 2012); wild deer in the USA and Canada (Nishi et al. 2006; O’Brien et al. 2011); buffalo and other wildlife in South Africa (Renwick et al. 2007) and European bison (Bison bonasus) and wild boar (Sus scrofa) in Europe (Naranjo et al. 2008; Gortázar et al. 2011; Krajewska et al. 2015). In an infected animal, M. bovis can be excreted in all bodily fluids, meaning transmission is possible directly through close contact (aerosol transmission, meat or milk ingestion) or indirectly via contact with fluids or excreta in the environment (Neill et al. 2001; Radostits et al. 2007).
Control of TB is particularly complicated where there are multiple livestock and wildlife hosts. This is the case in south-central Spain, an area where relatively low density cattle and pig (Sus scrofa domesticus) rearing occurs alongside red deer (Cervus elaphus) and wild boar hunting activities (Kukeilka et al. 2013; Cowie et al. 2014). There is therefore a significant potential for interactions between large game and domestic species, providing opportunities for disease transmission and maintenance. Bovine TB outbreaks occur in distinct clusters within the region and are significantly positively associated with wild boar TB prevalence and hunting activities (Rodríguez-Prieto et al. 2012; Martínez-López et al. 2013). The presence of wildlife has also been shown to be a significant risk factor for bovine TB at the individual farm level (Cowie et al. 2014). Both cattle and wildlife display high TB prevalence in south-central Spain, and levels in wild boar and red deer have increased over the last 12 years (Vicente et al. 2013). It is thought that the presence of these wildlife reservoirs is contributing to the failure to eradicate TB in the cattle population despite a long-standing test-and-slaughter scheme (Diez et al. 2002; Naranjo et al. 2008; Gortazar et al. 2011).
In this study, we aimed to spatially and temporally quantify direct and indirect interactions between cattle, pigs, red deer and wild boar in a mixed community infected with M. bovis. Using these data, we tested the hypothesis that direct interactions would be much less frequent than indirect ones and that the frequency of both types of interactions would be enhanced by food and water sites, due to the aggregation of individuals from multiple species at these locations. Furthermore, we hypothesised that seasonal variation in the availability of food and water sites would alter this pattern of aggregation (and hence the level of interactions) throughout the year. Finally, we investigated the frequency of indirect interactions that occurred within a range of critical time windows that relate to the environmental survival time of M. bovis and other diseases. To our knowledge, this is the first study to quantify close interactions between known individuals from multiple wildlife and livestock species.
Methods
Study area
The landscape in south-central Spain is characterised by the agroforestry system “dehesa”, an open savannah-like habitat with low densities of oak trees (Quercus spp), commonly adjacent to areas of forest and scrubland (Gaspar et al. 2009). Extensive livestock rearing of beef cattle, small ruminants (sheep and some goats) and Iberian pigs takes place here at low stocking densities (Plieninger et al. 2004; Milan et al. 2006; Gaspar et al. 2008). Farm sizes are highly variable, with a recent study identifying a mean size of 467 ha (range 37–2040 ha) (Cowie et al. 2014). The area has high densities of red deer and wild boar (Acevedo et al. 2008) where they are managed for recreational hunting activities, often on the same or adjacent land as livestock farming (Herruzo and Martinez-Jauregui 2013), where densities tend to be lower. TB is prevalent in this area despite test-and-slaughter schemes, affecting 3.54 % of cattle herds in the area in 2012 (RASVE 2014). Large-scale and long-term sampling studies have also revealed a high prevalence in wild boar (59.0 %) and red deer (9.4 %) in the region (Vicente et al. 2013).
Environmental conditions vary throughout the year, with temperature ranging from below 0 to over 40 °C. A wet season starts in autumn and typically contributes most of the annual rainfall within 3 months. As a result, water and food for animals are often limited in the peak of the dry season (from June to September) and livestock receive supplementary food and water from artificial sources (Kukeilka et al. 2013). At the end of the dry season, acorns fall from the oak trees (an event called “Montanera”) providing food for animals, especially pigs and wildlife species.
Study farm
This study was conducted on a regionally representative, traditionally managed beef cattle-rearing farm which also conducts hunting activities, located in Ciudad Real province. The farm covers 300 ha and was rearing an average of 90 “Retinta” breed cattle and 5 adult Iberian pigs during the study period. Cattle and pigs received supplementary food at two main locations in the summer months. Simple wire fencing was used at the farm boundaries. This contained cattle but allowed pigs and wildlife species to cross into and out of the farm. Adjoining properties were used for cattle and pig rearing and/or as hunting estates.
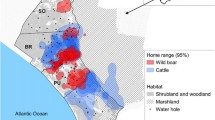
The farm is considered representative of cattle farms in the region because of typical management practices, stocking densities, environmental conditions and TB prevalence. One difference is that parts of the farm border the Montoro reservoir (Fig. 1), meaning that wildlife and livestock had greater access to water year round than is often available on other farms in the region. Within the farm, water was available from seasonal streams, managed by two small reservoirs.
Wildlife densities on the study farm are moderate compared with those on hunting estates in the area, but representative of cattle farms, with night-time spotlight transects revealing no more than 15–20 red deer, 10 roe deer and 10 wild boar on the 300 ha farm. Government sampling of the farm’s cattle with skin tests confirmed by culture showed a mean TB prevalence of 8.65 % during the study period. Wildlife shot on the farm and the neighbouring hunting estates were tested for M. bovis infection following methods used by Vicente et al. (2006). This revealed mean local prevalences of 84 % in wild boar and 30 % in red deer. Samples from the Iberian pigs on the farm were also tested, with seropositivity indicating that 36 % (n = 25, including juveniles) had antibodies against members of the M. tuberculosis complex (Aurtenetxe et al. 2008).
Data collection
We deployed proximity data loggers (Sirtrack Tracking Solutions, New Zealand) and GPS location loggers (Microsensory Systems, Spain) on the livestock and wildlife species, as well as at base stations around the farm. Prior to collaring any animals, base stations with proximity loggers were placed at the site of key resources on the farm—food points, water sources and control points (Fig. 1). Food points are places where concentrated cattle feed was provided, either in permanent raised troughs or on the ground. Control points were placed at random within areas with none of the key resources and no animal paths that led to any key resources. Base stations were set to record contacts at UHF30, triggering recording at an estimated radius of 3.1 m around the base station (Goodman 2007). Base stations were placed 1–2 m above the ground, hung from existing natural features such as dead trees where possible and were left in place for 1 month before recording began to avoid bias caused by animal investigation of the novel objects.
Cattle collaring took place during routine veterinary inspections of the cattle, whilst they were restrained in the farm’s own cattle race. Pigs were captured individually and held by hand during collaring. Collaring red deer and wild boar required the use of traps, following methods described in greater detail by Barasona et al. (2013a). The collaring strategy was therefore opportunistic and dependent on the animals captured. All animal proximity loggers were set to a UHF setting of 45, triggering recording if another collared animal comes within a radius of 1.5 m (Goodman 2007). This differed from the base stations in order to ensure that only very close contacts between animals were recorded as direct contacts. Battery life was estimated to be a maximum of 9 months so animals were re-collared within this time window where possible. Four GPS loggers were available, and these were deployed to maximise information from wildlife species (Barasona et al. 2014a). Where collars did not fall off, they were recovered during routine game hunting events.
Data processing
Following downloading of data from the collars, information from the 24-h periods immediately after collaring and immediately before collar retrieval was removed from the data set. Each dyad of animals that contacted each other should have two reciprocal data sets. In reality, these often differ slightly, so the longest data set for each dyad was retained for further analysis on the assumption that a contact may have been missed by one collar, but could not have been generated by the other collar if the contact had not occurred. One-way ANOVA tests were used to compare the means of the used and rejected data sets for each dyad.
The collars recorded two contacts with an interval between them of up to 17 s as one long continuous contact. This time duration is known as the separation time. Short separation times allow for fine scale data on direct contacts, but have been shown to increase the likelihood of broken contacts, where one long contact is recorded as multiple shorter contacts (Prange et al. 2006). These broken contacts are often of 1-s duration and are not considered valid contacts when evaluating possible disease transmission. Using Drewe et al.’s (2012) ‘contactweld’ function in R (version 2.15.3, R Core Development Team 2012) contacts with a separation time of 60 s or less were merged into single longer contacts. After this, any remaining 1-s contacts were removed from the data set.
Connectedness to resources
Raw contact data do not account for the amount of time each individual wore the logger and the number of base stations it could contact (which did vary slightly). The connectivity measures C freq and C dur were therefore calculated to allow for comparisons between base station types. These were calculated for each species, following the methods of Böhm et al. (2009), as:
Indirect interactions data analysis
GPS data for each species allowed for the calculation of home range areas and the overlaps between these home ranges. Using the ‘adehabitat’ package in R, the kernel density estimation was used to calculate the utilisation distribution (UD) with least-squares cross validation for each individual. This method does not rely on predetermined distributions, allows for the incorporation of various centres of activity and can be used where limited data are available. Core and home ranges were set as 50 and 95 % UD, respectively (Barasona et al. 2014a), and were calculated for each season. GPS data also provided information about the study animals’ movements, both on and off the farm. Using the spatial analysis programme QGIS (version 1.8.0, Quantum GIS Development Team 2013), fixes were classified as inside or outside the farm boundary, and the number of times each individual crossed the boundary was calculated and standardised by the number of days each animal was collared.
Finally, the intervals between visits to base stations by two individuals of the same or different species were classified into a range of critical time windows (CTWs). Thirteen CTWs were selected, ranging from 30 s to 12 days, to account for the known environmental survival times of different livestock diseases. The number and percentage of contacts and the number divided by the number of base stations at each resource were calculated for each dyad of species.
Results
Data returns
Proximity data loggers returned over 400,000 direct contacts between 17 cows, 8 pigs, 2 red deer and 2 wild boars. This represented 24, 63, 13 and 20 % of the estimated available population of each species, respectively. Data were collected continuously for 2 years starting in summer 2010, though there were only 202 consecutive days where all four species types were collared at the same time.
After removing reciprocal contacts from each dyad of individuals, significant levels of agreement between the used and rejected data sets were identified. ANOVA test p values ranged from 0.02 to <0.0001, with R 2 values from 0.61 to 0.97. The data contained large proportions of 1-s contacts. To provide an example with the cattle contact data, the raw data contained 43.1 % (95 % CI = 35.7–50.4) 1-s contacts. Merging contacts with a separation time of less than 60 s resulted in a mean of 50.3 % (95 % CI = 42.4–58.1) reduction in the number of these contacts. However, the percentage of 1-s contacts remained high after merging, at 47.0 % (95 % CI = 44.5–49.5). All remaining 1-s contacts were then removed from further analysis. Similar patterns were observed in data for all species and the same processes were applied, resulting in a direct contact data set of 57,188 contacts.
Proximity loggers at base stations returned over 75,000 raw contacts from four controls, two foods and ten water points around the farm (Fig. 1). As base stations were set to record at a different detection distance, the data were always taken from the base station records and reciprocal contacts on the collars were discarded. After merging contacts to a separation time of 60 s and removing 1-s contacts, 12,628 contacts remained for further analysis.
GPS loggers were attached to two cows, one pig, two red deer and two wild boars. The herd behaviour of the pigs and cattle means that the livestock GPS data normally represent the approximate position of the majority of the animals of that species. After the removal of any erroneous records, 43,595 fixes were used in further analysis. Most fixes were recorded from cattle (43 %) and red deer (44 %), whilst pigs (5 %) and wild boar (8 %) returned fewer fixes because they more frequently lost satellite reception and tended to lose their collars. All species recorded adequate fixes to exceed the minimum of 50 fixes required for kernel home range estimation (Seaman et al. 1999).
Direct interactions
Of the 57,188 direct contacts recorded, 875 (1.53 %) contacts were observed between different species and 216 (0.38 %) were between wildlife and livestock (Fig. 2). Variation was observed over the seasons for both contact frequency and mean contact duration. Cattle-pig and cattle-red deer contacts occurred more frequently in autumn and winter. Considerable variation was observed in the duration of contacts. Cattle-cattle and pig-pig contacts were each significantly longer in winter (GLM, both p < 0.001), and cattle-pig contacts had significantly longer durations in autumn (GLM, p < 0.001).
Network representation of the direct contacts observed between cattle, pigs, wild boar (WB) and red deer (RD) on the study farm. Each node represents an individual animal and the width of the lines between them is proportionate to the number of contacts observed. Nodes are arranged to visualise the social groups observed but the proximity of nodes to one another is illustrative and has no spatial relevance
Home range overlaps
Mean livestock home ranges (95%UD) were 2.08 km2 in cattle and 4.21 km2 in pigs. Mean home ranges for wildlife were 7.25 km2 in red deer and 1.96 km2 in wild boar. Home range overlaps were high, particularly between red deer and cattle, whose combined home ranges overlapped by over 4.0 km2 (27 and 95 % of the total range for each species, respectively) throughout the year. Overlaps between individual red deer, cattle and pigs, and pigs and red deer all peaked considerably in autumn. Core (50%UD) ranges were much smaller, with only red deer and cattle maintaining a core range overlap of over 0.5 km2 (3 and 12 %, respectively) throughout the year. Cattle-wild boar overlaps were comparatively low, with no core range overlap recorded.
Farm boundary crossings
The GPS data also allowed us to investigate animal movements across the farm boundary. Cattle were unable to cross farm boundaries, but pigs showed the highest rate of boundary crossings at 1.4 crossings per day. Red deer showed far more frequent crossings (1.19 crossings individual−1 day−1) than wild boar (0.61 crossings individual−1 day−1).
Daily activity patterns
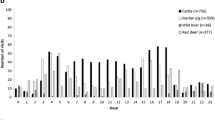
Similarities were observed in the daily activity patterns of red deer and cattle (Fig. 3), with both species most active at dawn and dusk. Other species did not show any notable alignment with cattle activity patterns.
Connectedness to resources
Over 50 % of all individuals of each species were detected at resource locations, with every individual except one red deer being detected at both food and water points. The same C freq and C dur measures of connectedness were calculated between each individual and each resource type. C freq (Fig. 4) varied considerably between species and resources. Livestock showed the highest connectedness to food locations. Wildlife appeared to use these resources much less, with wild boar showing some connection to control points. C dur measures (Fig. 5) displayed very similar patterns to C freq, showing that cattle are the only species to stay for long durations at resource points.
Critical time window analysis
The number of indirect interactions (where one animal visits a resource and then another of the same or a different species visits the same location at a later time) that occurred with intervals less than the range of CTWs were calculated for intra- (Fig. 6) and inter-species (Fig. 7) interactions. Most intra-species indirect interactions occurred within short CTWs, with 50 % or more occurring within 5 min at all resource types. When standardised by the number of base stations, the majority of these interactions occurred at food resource points. Pigs were also more likely to have indirect interactions at food sites, but these were more spread out, with 50 % of these interactions occurring within a 3-h interval. No indirect interactions were recorded between red deer at food sites, but 50 % of indirect interactions at water points fell within the 5-min CTW. Few indirect interactions between different species were observed, and most interactions fell within longer CTWs.
The standardised, cumulative number of intra-species indirect interactions per base station that fell within each critical time window at each resource type for a cattle, b pigs and c red deer. Legends on each graph show the total number of indirect interactions recorded during the study period. No data were available for wild boar as individuals were not collared concurrently. Where resource types are missing no indirect interactions were recorded at these locations. Note the different scales on each y-axis
The standardised, cumulative number of inter-species indirect interactions per base station that fell within each critical time window at each resource type for a cattle-pigs, b cattle-red deer and c cattle-wild boar. Legends on each graph show the total number of indirect interactions recorded during the study period. Where resource types are missing no indirect interactions were recorded at these locations. Note the different scales on each y-axis
Discussion
Data evaluation
Inter- and intra-species interactions recorded by proximity loggers cannot be interpreted directly as actual disease transmission opportunities, since they provide no information on the type of interaction that occurred. Nevertheless, the combination of GPS and proximity logger technology provides the best available spatially and temporally explicit data on local animal movements and social contacts (e.g. Drewe et al. 2012), and hence can serve as an indication of potential transmission opportunities. A larger sample of wildlife individuals would allow for more confidence in our conclusions. Problems were encountered with keeping collars on wild boar and pigs, attracting deer into traps, and with pigs occupying traps intended for wild boar. Wildlife were found at only moderate densities on the farm (Acevedo et al. 2007; Acevedo et al. 2008), which limited trapping success. However, the choice of study site was considered important as it is a representative of the study area. Data from proximity loggers has great potential for informing animal social networks and models (e.g. Marsh et al. 2011) but for detailed analysis, it is important to include the majority of the study population in order to draw confident conclusions. Furthermore, if social interactions are being studied in relation to disease transmission and the disease is known to survive in the environment, it is also very important to investigate when and where indirect interactions occur (Kukeilka et al. 2013).
Direct interactions
This study has quantified direct and indirect interactions between individuals in a multi-host disease community. As with similar research (Drewe et al. 2013), data on direct interactions between different species were limited, despite the long-term continuous sampling. Direct intra-species contacts were frequent, but direct inter-species interactions were relatively rare, identifying more opportunity for disease transmission within species than between species. Nonetheless, this highlights the possible importance of the environmental survival of pathogens and indirect interactions between individuals in maintaining disease.
The direct contacts that were recorded do indicate that cattle contacts with both red deer and pigs are most frequent in autumn (at the end of the dry season, during the acorn mast). Furthermore, cattle-pig contacts were significantly longer at this time. This relates to previous work on the study farm that showed the number of visits to farm resources increased through the dry season and peaked during the acorn season in autumn (Kukeilka et al. 2013), giving further confidence that the collars were identifying real changes in behaviour.
Indirect interactions
In our data set, year-round core and home range overlaps were highest for cattle and red deer, with all overlaps increasing in autumn (during the acorn mast). The acorn food source is the likely reason for aggregation of both species, and foraging in the same locations may increase the likelihood of transmission of pathogens between different individuals. Furthermore, foraging may be taking place at similar times for cattle and red deer as their daily activity levels follow such similar patterns. Red deer also crossed the farm boundary more than one time per day on average, increasing the risk of encountering disease outside the farm and then transmitting it to livestock within the farm. However, pigs also showed moderate levels of home range overlap with cattle and the highest rate of boundary crossings. Pigs may be an important part of the disease cycle in this region, as has been previously identified in another Mediterranean system in Sicily (Di Marco et al. 2012). Further research and testing of pig and red deer movements and disease in this area may be important for a fuller understanding of disease maintenance and spread. In terms of disease transmission, the relative importance of each species will depend on the dyad of species and the pathogen. For instance, red deer share more viral pathogens with cattle, whilst wild boars have more pathogens in common with pigs (Miller et al. 2013).
Most indirect interactions between individuals from different species occurred at water points. Red deer and cattle also showed the highest connectivity to water, though red deer generally stayed at water points for shorter durations. Water points should therefore be considered a potential hotspot for disease transmission between wildlife and livestock in south-central Spain. Indeed, Vicente et al. (2007) described a positive association between wildlife aggregation at water- and ground-level feeding sites and tuberculous-like lesions in animals from large game hunting estates, suggesting that host aggregation could drive disease transmission. Additionally, the density of water points on farms has been shown to be negatively associated with TB risk in cattle (Kaneene et al. 2002; Cowie et al. 2014) and in wildlife (Barasona et al. 2014b). This means that having fewer sources of water is associated with an increased risk of bovine TB on the farm. The availability of water on our study farm from the reservoir introduces potential bias into our results. However, as water points caused aggregation even with this other water source available, this suggests that water may be a very important risk factor for disease in places where it is more limited. In combination with the known TB prevalence data in the area and the knowledge that wildlife here support the disease in the absence of cattle (Gortazar et al. 2005; Corner 2006; Vicente et al. 2006; Naranjo et al. 2008), it seems likely that indirect interactions at key resource points are more important than direct contact in the transmission of TB in this area.
Critical time windows
Intra-species indirect interactions occurred much more frequently in short CTWs than inter-species interactions. Cattle and pigs had most of these contacts at supplementary food sites, though red deer never utilised the same food sites and instead showed most indirect interactions at water points. All species showed more indirect interactions at resource sites compared to control sites, indicating aggregation occurring at these resources. However, indirect inter-species interactions were still avoided, with less than 50 % of use of the same resources by different species occurring within a 6-h CTW. Indirect interactions between cattle and pigs or wild boar happened mostly at food sites, whereas red deer used the same water resources. This may be because of differences in foraging strategy; it may only be worthwhile for suids, who will forage by rooting, to investigate a food site after cattle have consumed the majority of the food.
Regarding TB, a review of the literature on the survival of M. bovis in the environment applied to this region suggests that on average, there could be a 3-day CTW in the dry season and a 12-day CTW in the wet season (Kukeilka et al. 2013). Over 90 % of intra-species indirect interactions at food or water resources occurred within a 3-day CTW, suggesting that if a pathogen is present within a population, this could be important in the maintenance and spread of the disease. Between different species, we see 40–66 % of indirect interactions occurring within 3 days, rising to 72–100 % within the 12-day wet season CTW. In the same region, Barasona et al. (2013b) demonstrated that separating wildlife and livestock access to water resources appears to have reduced TB prevalence in cattle. As pigs and cattle were more closely connected to food resources, it may also be important to consider separation of livestock at feeding stations. This is currently especially important in the study region as pigs are not subject to specific controls for TB (RASVE 2014).
Management implications
As most direct and indirect interactions occur within species, livestock disease testing and management (such as test-and-slaughter schemes) remain important as a means of controlling TB. However, enough direct and indirect interactions between livestock and wildlife were observed to indicate that multiple host diseases need to be managed in the context of the whole disease community. Pigs were well connected within the indirect contact network of the host community through shared use of food and water resources. This is consistent with the home range overlaps with cattle that peaked during the autumn acorn mast. At present, pigs are not monitored for TB in Europe, though M. bovis infection is increasingly reported in pigs in several countries (e.g. Di Marco et al. 2012; Bailey et al. 2013). In south-central Spain, pigs are increasingly being refused by slaughterhouses due to visible TB-compatible lesions (Gomez-Laguna et al. 2010). Furthermore, the TB seroprevalence of over 35% in the pigs in this study suggests further research into the significance of pigs as a TB host would improve our understanding of the TB system in this region.
Thus far, research efforts in this area have often focussed on wild boar as the main wildlife disease reservoir. However, in this study, cattle showed more direct and indirect interactions with red deer than with wild boar. Our data showed that red deer cross farm boundaries frequently, moving into hunting estates and/or other livestock farms. Within the farm, they had large home range overlaps with cattle year round, with direct contacts with cattle occurring significantly more often in autumn and winter. Furthermore, their daily activity patterns were notably similar to those of cattle, particularly in the morning. These results suggest that the possible role of red deer in the disease system in this region of Spain should receive further investigation, though the pathogenesis and excretion of a pathogen by each species need to be considered. Regarding TB, red deer prevalence in the region has been shown to be stable around a mean of 9.4 %, whilst mean wild boar TB prevalence is 59.0 % and has been increasing over the last decade (Vicente et al. 2013). Further research into direct and indirect interactions between wild boar and livestock is also necessary.
Though limited, direct inter-species interactions may still have epidemiological significance. However, indirect interactions not only occurred more frequently than direct interactions but did so within short critical time windows. Understanding the environmental survival time of a minimum infective dose of a disease would show which of the CTWs proposed in this study were likely to be important. Regarding TB, a recent literature review identified only 15 studies published between 1930 and 2011 that estimated M. bovis environmental survival times. Survival times ranged from no M. bovis recovery to recovery of genetic material after 300 days, on a wide range of substrates. In our study, intra-species indirect interactions occurred within short CTWs, suggesting they would also be a potential route for infection with viral diseases which tend to have shorter environmental survival times (Kukeilka et al. 2013). The role of indirect interactions in disease transmission within and between species needs to be explored further.
Conclusions
This study has contributed to our understanding of multi-species host communities, quantifying direct and indirect interactions between multiple species and showing how they are influenced by the seasonal and spatial distribution of key resources. Management strategies that reduce shared wildlife and livestock space use are likely to achieve greater reductions in inter-species disease transmission than if this is not considered. The efficiency of these strategies could be enhanced by concentrating effort during seasons when low water availability or high food availability causes aggregation of multiple species.
References
Acevedo P, Vicente J, Höfle U et al (2007) Estimation of European wild boar relative abundance and aggregation: a novel method in epidemiological risk assessment. Epidemiol Infect 135:519–527. doi:10.1017/S0950268806007059
Acevedo P, Ruiz-Fons F, Vicente J et al (2008) Estimating red deer abundance in a wide range of management situations in Mediterranean habitats. J Zool 276:37–47. doi:10.1111/j.1469-7998.2008.00464.x
Anderson RM, May RM (1979) Population biology of infectious diseases: part I. Nature 280:361–367. doi:10.1038/280361a0
Anderson DP, Ramsey DSL, Nugent G et al (2013) A novel approach to assess the probability of disease eradication from a wild-animal reservoir host. Epidemiol Infect 141:1509–1521. doi:10.1017/S095026881200310X
Aurtenetxe O, Barral M, Vicente J et al (2008) Development and validation of an enzyme-linked immunosorbent assay for antibodies against Mycobacterium bovis in European wild boar. BMC Vet Res 4:43. doi:10.1186/1746-6148-4-43
Bailey SS, Crawshaw TR, Smith NH, Palgrave CJ (2013) Mycobacterium bovis infection in domestic pigs in Great Britain
Barasona JA, López-Olvera JR, Beltrán-Beck B et al (2013a) Trap-effectiveness and response to tiletamine-zolazepam and medetomidine anaesthesia in Eurasian wild boar captured with cage and corral traps. BMC Vet Res 9:107. doi:10.1186/1746-6148-9-107
Barasona JA, VerCauteren KC, Saklou N et al (2013b) Effectiveness of cattle operated bump gates and exclusion fences in preventing ungulate multi-host sanitary interaction. Prev Vet Med 111:42–50. doi:10.1016/j.prevetmed.2013.03.009
Barasona JA, Latham M, Acevedo P et al (2014a) Spatiotemporal interactions between wild boar and cattle: implications for cross-species disease transmission. Vet Res 45:122. doi:10.1186/s13567-014-0122-7
Barasona JA, Mulero-Pázmány M, Acevedo P et al (2014b) Unmanned aircraft systems for studying spatial abundance of ungulates: relevance to spatial epidemiology. PLoS One 9:e115608. doi:10.1371/journal.pone.0115608
Böhm M, Palphramand KL, Newton-Cross G et al (2008) Dynamic interactions among badgers: implications for sociality and disease transmission. J Anim Ecol 77:735–745. doi:10.1111/j.1365-2656.2008.01377.x
Böhm M, Hutchings M, White P (2009) Contact networks in a wildlife-livestock host community: identifying high-risk individuals in the transmission of bovine TB among badgers and cattle
Cleaveland S, Laurenson MK, Taylor LH (2001) Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci 356:991–999. doi:10.1098/rstb.2001.0889
Cooper SM, Scott HM, de la Garza GR et al (2010) Distribution and interspecies contact of feral swine and cattle on rangeland in south Texas: implications for disease transmission. J Wildl Dis 46:152–164. doi:10.7589/0090-3558-46.1.152
Corner LAL (2006) The role of wild animal populations in the epidemiology of tuberculosis in domestic animals: how to assess the risk. Vet Microbiol 112:303–312. doi:10.1016/j.vetmic.2005.11.015
Cowie CE, Beck BB, Gortazar C et al (2014) Risk factors for the detected presence of Mycobacterium bovis in cattle in south central Spain. Eur J Wildl Res 60:113–123. doi:10.1007/s10344-013-0757-0
Craft ME, Volz E, Packer C, Meyers LA (2011) Disease transmission in territorial populations: the small-world network of Serengeti lions. J R Soc Interface 8:776–786. doi:10.1098/rsif.2010.0511
Di Marco V, Mazzone P, Capucchio MT et al (2012) Epidemiological significance of the domestic black pig (Sus scrofa) in maintenance of bovine tuberculosis in Sicily. J Clin Microbiol 50:1209–1218. doi:10.1128/JCM.06544-11
Diez M, Huerta C, Moreno T et al (2002) Tuberculosis in Spain: epidemiological pattern and clinical practice. Int J Tuberc Lung Dis 6:295–300
Drewe JA (2010) Who infects whom? Social networks and tuberculosis transmission in wild meerkats. Proc R Soc B Biol Sci 277:633–642. doi:10.1098/rspb.2009.1775
Drewe JA, Weber N, Carter SP et al (2012) Performance of proximity loggers in recording intra- and inter-species interactions: a laboratory and field-based validation study. PLoS One 7:e39068. doi:10.1371/journal.pone.0039068
Drewe JA, O’Connor HM, Weber N et al (2013) Patterns of direct and indirect contact between cattle and badgers naturally infected with tuberculosis. Epidemiol Infect 141:1–9. doi:10.1017/S0950268813000691
Fitzgerald SD, Kaneene JB (2013) Wildlife reservoirs of bovine tuberculosis worldwide: hosts, pathology, surveillance, and control. Vet Pathol 50:488–499. doi:10.1177/0300985812467472
Gaspar P, Escribano M, Mesías FJ et al (2008) Sheep farms in the Spanish rangelands (Dehesas): typologies according to livestock management and economic indicators. Small Rumin Res 74:52–63. doi:10.1016/j.smallrumres.2007.03.013
Gaspar P, Mesı FJ, Escribano M, Pulido F (2009) Sustainability in Spanish extensive farms (Dehesas): an economic and management indicator-based evaluation. Rangel Ecol Manag 62:153–162
Gomez-Laguna J, Carrasco L, Ramis G et al (2010) Use of real-time and polymerase chain reaction assays for the diagnosis of porcine tuberculosis in formulin-fixed, paraffin-embedded tissues. J Vet Diagn Investig 22:123–128
Goodman E (2007) Quantifying interactions in a high-density badger (Meles meles) population. University of York
Gortazar C, Vicente J, Samper S et al (2005) Molecular characterization of Mycobacterium tuberculosis complex isolates from wild ungulates in south-central Spain. Vet Res 36:43–52. doi:10.1051/vetres
Gortázar C, Delahay RJ, Mcdonald RA et al (2011) The status of tuberculosis in European wild mammals. Mamm Rev 42:193–206. doi:10.1111/j.1365-2907.2011.00191.x
Gortazar C, Vicente J, Boadella M et al (2011) Progress in the control of bovine tuberculosis in Spanish wildlife. Vet Microbiol 151:170–178. doi:10.1016/j.vetmic.2011.02.041
Haydon DT, Cleaveland S, Taylor LH, Laurenson MK (2002) Identifying reservoirs of infection: a conceptual and practical challenge. Emerg Infect Dis 8:1468–1473
Herruzo AC, Martinez-Jauregui M (2013) Trends in hunters, hunting grounds and big game harvest in Spain. For Syst 22:114. doi:10.5424/fs/2013221-03371
Hutchings MR, Harris S (1997) Effects of farm management practices on cattle grazing behaviour and the potential for transmission of bovine tuberculosis from badgers to cattle. Vet J 153:149–162. doi:10.1016/S1090-0233(97)80035-4
Hutchings MR, Harris S (1999) Quantifying the risks of TB infection to cattle posed by badger excreta. Epidemiol Infect 122:167–173
Kaneene JB, Bruning-Fann C, Granger L et al (2002) Environmental and farm management factors associated with tuberculosis on cattle farms in northeastern Michigan. J Am Vet Med Assoc 221:837–842
Krajewska M, Zabost A, Welz M et al (2015) Transmission of Mycobacterium caprae in a herd of European bison in the Bieszczady Mountains, Southern Poland. Eur J Wildl Res 61:429–433. doi:10.1007/s10344-015-0912-x
Kukeilka E, Barasona JA, Cowie CE et al (2013) Spatial and temporal interactions between livestock and wildlife in South Central Spain. Prev Vet Med 112:213–221
Marsh MK, McLeod SR, Hutchings MR, White PCL (2011) Use of proximity loggers and network analysis to quantify social interactions in free-ranging wild rabbit populations. Wildl Res 38:1–12. doi:10.1071/WR10150
Martin C, Pastoret P-P, Brochier B et al (2011) A survey of the transmission of infectious diseases/infections between wild and domestic ungulates in Europe. Vet Res 42:70. doi:10.1186/1297-9716-42-70
Martínez-López B, Barasona JA, Gortázar C et al (2013) Farm-level risk factors for the occurrence, new infection or persistence of tuberculosis in cattle herds from South-Central Spain. Prev Vet Med. doi:10.1016/j.prevetmed.2013.11.002
Michel AL, Müller B, van Helden PD (2010) Mycobacterium bovis at the animal-human interface: a problem, or not? Vet Microbiol 140:371–381. doi:10.1016/j.vetmic.2009.08.029
Miguel E, Grosbois V, Caron A et al (2013) Contacts and foot and mouth disease transmission from wild to domestic bovines in Africa. Ecosphere 4:1–32. doi:10.1890/es12-00239.1
Milan M, Bartolome J, Quintanilla R et al (2006) Structural characterisation and typology of beef cattle farms of Spanish wooded rangelands (Dehesas). Livest Sci 99:197–209
Miller RS, Farnsworth ML, Malmberg JL (2013) Diseases at the livestock-wildlife interface: status, challenges, and opportunities in the United States. Prev Vet Med 110:119–132. doi:10.1016/j.prevetmed.2012.11.021
Naranjo V, Gortazar C, Vicente J, de la Fuente J (2008) Evidence of the role of European wild boar as a reservoir of Mycobacterium tuberculosis complex. Vet Microbiol 127:1–9. doi:10.1016/j.vetmic.2007.10.002
Neill SD, Bryson DG, Pollock JM (2001) Pathogenesis of tuberculosis in cattle. Tuberculosis (Edinb) 81:79–86. doi:10.1054/tube.2000.0279
Nishi JS, Shury T, Elkin BT (2006) Wildlife reservoirs for bovine tuberculosis (Mycobacterium bovis) in Canada: strategies for management and research. Vet Microbiol 112:325–338. doi:10.1016/j.vetmic.2005.11.013
O’Brien DJ, Schmitt SM, Fitzgerald SD, Berry DE (2011) Management of bovine tuberculosis in Michigan wildlife: current status and near term prospects. Vet Microbiol 151:179–187. doi:10.1016/j.vetmic.2011.02.042
O’Connor CM, Haydon DT, Kao RR (2012) An ecological and comparative perspective on the control of bovine tuberculosis in Great Britain and the Republic of Ireland. Prev Vet Med 104:185–197. doi:10.1016/j.prevetmed.2011.11.010
Plieninger T, Pulido FJ, Schaich H (2004) Effects of land-use and landscape structure on holm oak recruitment and regeneration at farm level in Quercus ilex L. dehesas. J Arid Environ 57:345–364. doi:10.1016/S0140-1963(03)00103-4
Prange S, Jordan T, Hunter C, Gehrt SD (2006) New radiocollars for the detection of proximity among Individuals. Wildl Soc Bull 34:1333–1344. doi:10.2193/0091-7648(2006)34[1333:NRFTDO]2.0.CO;2
Quantum GIS Development Team (2013) Quantum GIS geographic information system
Radostits OM, Gay CC, Hinchcliff KW, Constable PD (2007) Veterinary medicine: a textbook of the diseases of cattle, horses, sheep, pigs and goats. Springer, New York
RASVE (2014) Informe final téchnico-financiero programa nacional de la tuberculosis bovine año 2012
R Core Development Team (2012) A language and environment for statistical computing
Renwick AR, White PCL, Bengis RG (2007) Bovine tuberculosis in southern African wildlife: a multi-species host-pathogen system. Epidemiol Infect 135:529–540. doi:10.1017/S0950268806007205
Rodríguez-Prieto V, Martínez-López B, Barasona JA et al (2012) A Bayesian approach to study the risk variables for tuberculosis occurrence in domestic and wild ungulates in South Central Spain. BMC Vet Res 8:148. doi:10.1186/1746-6148-8-148
Schiller I, Waters WR, Vordermeier HM et al (2011) Bovine tuberculosis in Europe from the perspective of an officially tuberculosis free country: trade, surveillance and diagnostics. Vet Microbiol 151:153–159. doi:10.1016/j.vetmic.2011.02.039
Seaman DE, Millspaugh JJ, Kernohan BJ et al (1999) Effects of sample size on kernel home range estimates. J Wildl Dis 63:739–747
Vander Wal E, Paquet PC, Andrés JA (2012) Influence of landscape and social interactions on transmission of disease in a social cervid. Mol Ecol 21:1271–1282. doi:10.1111/j.1365-294X.2011.05431.x
Vicente J, Hofle U, Garrido J et al (2006) Wild boar and red deer display high prevalences of tuberculosis-like lesions in Spain. Vet Res 37:107–119. doi:10.1051/vetres
Vicente J, Delahay RJ, Walker NJ, Cheeseman CL (2007) Social organization and movement influence the incidence of bovine tuberculosis in an undisturbed high-density badger Meles meles population. J Anim Ecol 76:348–360. doi:10.1111/j.1365-2656.2006.01199.x
Vicente J, Barasona JA, Acevedo P et al (2013) Temporal trend of tuberculosis in wild ungulates from Mediterranean Spain. Transbound Emerg Dis 60(Suppl 1):92–103. doi:10.1111/tbed.12167
Walrath R (2011) Efficacy of proximity loggers for detection of contacts between maternal pairs of white‐tailed deer. Wildl Soc Bull 35:452–460
zu Dohna H, Peck DE, Johnson BK et al (2014) Wildlife-livestock interactions in a western rangeland setting: quantifying disease-relevant contacts. Prev Vet Med 113:447–456. doi:10.1016/j.prevetmed.2013.12.004
Acknowledgments
We would like to thank the many research assistants and volunteers who contributed to this project—particularly Ignacio Cristóbal who helped maintain the equipment and study site. Esteban Arias made the project possible by allowing us to access his farm and animals. Francisco the farm hand provided invaluable support in cattle collaring and wildlife trapping. SRUC and IREC provided access to equipment and vehicles. SRUC receives support from the Scottish Government. C.E.C. was supported by an NERC/ESRC Interdisciplinary studentship. This is a contribution to PlanNacional I + D + i research grant AGL2011-30041 and AGL2014-56305 from MINECO and the EU FP7 grant WildTBvac (project number 613779). Funding was also received from ‘ASFORCE’, European Union’s Seventh Framework Programme (ref. FP7 - KBBE.2012.1.3-02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Study procedures were approved by the Animal Experiment Committee of Castilla-La Mancha University and were designed and developed by scientists (B and C animal experimentation categories) approved by the Spanish Ethic Committee. All applicable international, national and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Cowie, C.E., Hutchings, M.R., Barasona, J.A. et al. Interactions between four species in a complex wildlife: livestock disease community: implications for Mycobacterium bovis maintenance and transmission. Eur J Wildl Res 62, 51–64 (2016). https://doi.org/10.1007/s10344-015-0973-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10344-015-0973-x