Abstract
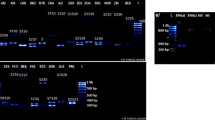
In this study, 48 almond (Prunus dulcis Miller [D.A. Webb]) genotypes were analyzed in terms of detailed morphological parameters. On the other hand, the incompatibility S genotypes in these genotypes were determined using a polymerase chain reaction (PCR) approach with allele-specific primers. High morphological diversity among the genotypes was observed. Most of the total variation (71.94%) in the seven phenological and five morphological traits consisted of the first three main principal components (PCs). The all-important traits ensured a positive value, but leaf colour had a negative value for the first three PCs. On the other hand, foliation time and petiole length showed low variation among the studied genotypes. According to the results of PCR using the AS1II- and AmyC5R-specific primers in a single reaction, the amplification was successful. The results showed amplification of nine different self-incompatibility alleles (S1, S2, S3, S5, S6, S10, S11, S12 and S13) and of the self-compatibility allele Sf. The PCR-amplified fragments ranged from 600 to 1600 bp. The self-compatibility allele Sf and S3 allele had the same band size at 1200 bp. The number of self-compatible genotypes was 12, including ‘Marta’, ‘F. Barese’, ‘Tuono’ and ‘Super Nova’ cultivars. S1, S2, S5 and S6 were the most common alleles, as each was found in almond genotypes assayed here. The least common alleles were S10, S11, S12 and S13 alleles, and especially S10 was determined only in ‘Dokuzoguz’ cultivar. The PCR approach is an easy, low-cost tool for early identification of self-compatible progeny seedlings. From these results, it could be concluded that these local genotypes might be considered as potential candidates to be used in breeding programs.



Similar content being viewed by others
References
Alonso JM, Socias-Company R (2006) Almond S‑genotype identification by PCR using specific and non-specific allele primers. Acta Hortic 726:67–70
Aslantas R, Guleryuz M (1999) Almond selection in microclimate areas of northeast Anatolia. XI Grempa Seminar on Pistacios and Almonds, Sanliurfa, 01–04 September 1999, pp 339–342
Boskovic R, Tobutt KR, Batlle I, Duval H, Martinez-Gomez P, Gradziel TM (2003) Stylar ribonucleases in almond: Correlation with, and prediction of incompatibility genotypes. Plant Breed 122:70–76
Channuntapipat C, Sedgley M, Collins G (2001) Sequences of the cDNAs and genomic DNAs encoding the S1, S7, S8, and Sf alleles from almond, Prunus dulcis. Theor Appl Genet 103:1115–1122
Channuntapipat C, Wirthensohn M, Ramesh SA, Battle I, Arus P, Sedgley M, Collins G (2003) Identification of incompatibility genotypes in almond (Prunus dulcis Mill.) using specific primers basen on the introns of the S‑alleles. Plant Breed 122:164–168
Colic S, Rakonjac V, Zec G, Nikolic D, Aksic MF (2012) Morphological and biochemical evaluation of selected almond [Prunus dulcis (Mill.) D.A.Webb] genotypes in northern Serbia. Turk J Agric For 36:429–438
De Giorgio D, Polignano GB (2001) Evaluating the biodiversity of almond cultivars from germplasm collection field in Southern Italy. Sustain Glob Farm 56:305–311
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Ercisli S (2004) A short review of the fruit germplasm resources of Turkey. Genet Resour Crop Evol 51:419–435
FAO (2021) Food and Agriculture Organization of the United Nations. “Crops data”. https://www.fao.org/faostat/en/#data/QCL
Hajilou J, Grigorian V, Mohammadi SA, Nazemieh A, Romero C, Vilanova S, Burgos L (2006) Self- and Cross- (in) compatibility between important apricot cultivars in northwest Iran. J Hortic Sci Biotechnol 81:513–517
Halasz J, Hegedus A, Pedryc A (2006) Review of the molecular background of selfincompatibility in rosaceous fruit trees. Int J Hortic Sci 12(2):7–18
Hanci F (2019) Genetic variability in peas (Pisum sativum L.) from Turkey asssessed with molecular and morphological markers. Folia Hort 31(1):101–116
Imani A, Amani G, Shamili M, Mousavi A, Rezai H, Rasouli M, Martínez-García PJ (2021) Diversity and broad sense heritability of phenotypic characteristic in almond cultivars and genotypes. Int J Hortic Sci Technol 8(3):281–289
Kester DE, Gradiziel TM (1996) Almonds. In: Janick J, Moore JN (eds) Nuts. Fruit Breeding, vol III. John Wiley & Sons, New York, pp 1–97
Khadivi-Khub A, Etemadi-Khah A (2015) Phenotypic diversity and relationships between morphological traits in selected almond (Prunus amygdalus) germplasm. Agroforest Syst 89:205–216
Lopez M, Vargas FJ, Batlle I (2006) Self-(in)compatibility almond genotypes: a review. Euphytica 150:1–16
Ma R, Oliveira MM (2001) Molecular cloning of the self-incompatibility genes S1 and S3 from almond (Prunus dulcis cv. Ferragnès). Sex Plant Reprod 14:163–167
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Martínez-Gómez P, López M, Alonso JM, Ortega E, Batlle I, Socías-Company R, Dicenta F, Dandekar AM, Gradziel TM (2003) Identification of self-incompatibility alleles in almond and related Prunus species using PCR. Acta Hortic 622:397–401
Mousavi A, Fatahi Moghadam MR, Zamani Z, Imani A (2010) Evaluation of quantitative and qualitative characteristics of some almond cultivars and genotypes. Iran J Hortic Sci 41(2):119–131
Mousavi A, Fatahi R, Zamani Z, Imani A, Dicenta F, Ortega E (2014) Genetic variation and frequency of S‑alleles in Iranian almond cultivars. Acta Hortic 1028:45–48
Nikoumanesh K, Ebadi A, Zeinalabedini M, Gogorcena Y (2011) Morphological and molecular variability in some Iranian almond genotypes and related Prunus species and their potentials for rootstock breeding. Sci Hortic 129:108–118
Odong TL, Van Heerwaarden J, Jansen J, Van Hintum TJL, Van Eeuwijk FA (2011) Determination of genetic structure of germplasm collections: are traditional hierarchical clustering methods appropriate for molecular marker data? Theor Appl Genet 123:195–205
Ortega E, Sutherland BG, Dicenta F, Boskovic R, Tobutt KR (2005) Determination of incompatibility genotypes in almond using first and second intron consensus primers: detection of new S alleles and correction of reported S genotypes. Plant Breed 124:188–196
Pinar H, Bircan M, Uzun A, Coskun OF, Yaman M (2016) Evulation of phenological and pomological characters of some almond genotypes and cultivars in Turkey. International Conference on Sustainable Development, Skopje, 19–23 September 2016, pp 385–388
Rasouli M (2017) The study of morphological traits and identification of self-incompatibility alleles in almond cultivars and genotypes. J Nuts 8(2):137–150
Rohlf FJ (2004) NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2.11V. Exeter software, Setauket, New York
Sakar H, El-Yamani M, Boussakouran A, Rharrabti Y (2019) Reproductive phenology of some local almond [Prunus dulcis (Mill.) D.A. Webb] genotypes from northern Morocco. J Anal Sci Appl Biotechnol 1(2):49–54
Sanchez-Perez R, Dicenta F, Martinez-Gomez P (2004) Identification of S‑alleles in almond using multiplex PCR. Euphytica 138:263–269
Sepahvand E, Khadivi-Khub A, Momenpour A, Fallahi E (2015) Evaluation of an almond collection using morphological variables to choose superior trees. Fruits 70(1):53–59
Sheikh-Alian A, Vezvaei A, Ebadi A, Fatahi-Moghadam MR, Sarkhosh A (2010) Determination and identification of selfincompatibility alleles in selective Iranian and foreign almond (Prunus dulcis M.) cultivars by PCR method. Iran J Hortic Sci 41(3):247–252
Sorkheh K, Shiran B, Rouhi V, Asadi E, Jahanbazi H, Moradi H, Gradziel TM, Martinez-Gómez P (2009) Phenotypic diversity within native Iranian almond (Prunus spp.) species and their breeding potential. Genet Resour Crop Evol 56:947–961
Sumbul A, Bayazit S (2019) Pomological and chemical attributes of almond genotypes from Hatay province (in Turkish). Int J Agric Wildl Sci 5(1):1–10
Talhouk SN, Lubani RT, Baalbaki R, Zurayk R, Al-Khatib A, Parmaksizian L, Jaradat AA (2000) Phenotypic diversity and morphological characterization of Amygdalus species in Lebanon. Genet Resour Crop Evol 47:93–104
Tamura M, Ushijima K, Sassa H, Hirano H, Tao R, Gradziel TM, Dandekar AM (2000) Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor Appl Genet 101:344–349
Valizadeh B, Ershadi A (2009) Identification of self-incompatibility alleles in Iranian almond cultivars by PCR using consensus and allele-specific primers. J Hortic Sci Biotechnol 84(3):285–290
Van Hintum TJL, Brown AHD, Spillane C, Hodgkin T (2000) Core collections of plant genetic resources. IPGRI Technical Bulletin No. 3,. nternational Plant Genetic Resources Institute, Rome
Vilanova S, Romero C, Llacer G, Badenes ML (2005) Identification of self incompatibility aleles in apricot by PCR and sequence analysis. J Am Soc Hortic Sci 130(6):893–898
Wirthensohn M, Rahemi M, Fernández-Martí A (2011) Identification of self-incompatibility genotypes and DNA fingerpriting of some Australian almond cultivars. Acta Hortic 912:561–566
Wunsch A, Hormoze JI (2004) S‑allele identification by PCR analysis in sweet cherry cultivars. Plant Breed 123:1–6
Yamane H, Tao R, Sugiora A (1999) Identification and cDNA cloning for S‑RNases in selfincompatible Japanese plum (Prunus salisina cv, sordum). Plant Biotechnol 16(5):389–396
Zeinalabedini M, Khayam-Nekoui M, Imani A, Majidian P (2012) Identification of selfcompatibility and self-incompatibility genotypes in almond and some Prunus species using molecular markers. Seed Plant Improv J 28(2):227–238
Funding
This work has been supported by Erciyes University Scientific Research Projects Coordination Unit under Grant number of FBA-2019-8708.
Author information
Authors and Affiliations
Contributions
Hasan Pınar contributed 35%, Ercan Yıldız 30%, Aydın Uzun 25%, Mustafa Bircan 10%.
Corresponding author
Ethics declarations
Conflict of interest
H. Pınar, E. Yıldız, M. Bircan and A. Uzun declare that they have no competing interests.
Additional information
Availability of data and material
Available
Rights and permissions
Springer Nature oder sein Lizenzgeber (z.B. eine Gesellschaft oder ein*e andere*r Vertragspartner*in) hält die ausschließlichen Nutzungsrechte an diesem Artikel kraft eines Verlagsvertrags mit dem/den Autor*in(nen) oder anderen Rechteinhaber*in(nen); die Selbstarchivierung der akzeptierten Manuskriptversion dieses Artikels durch Autor*in(nen) unterliegt ausschließlich den Bedingungen dieses Verlagsvertrags und dem geltenden Recht.
About this article
Cite this article
Pınar, H., Yıldız, E., Bircan, M. et al. Identification of Genetic Diversity Using Morphological Properties and Self-Incompatibility Alleles in Selected Prunus dulcis Miller (D.A. Webb) Genotypes. Erwerbs-Obstbau 65, 1595–1602 (2023). https://doi.org/10.1007/s10341-023-00874-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10341-023-00874-z