Abstract
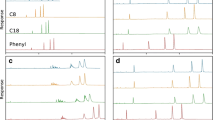
The differences in chromatographic behaviour of individual deoxynucleotides as well as small single-stranded and double-stranded DNA molecules have been examined for two resins from the Capto family: Capto Adhere and Capto Q ImpRes. Capto Adhere carries a multimodal ligand which combines strong anion with aromatic recognition, while Capto Q ImpRes is a strong anion exchanger with a chemically similar ligand, but without a phenyl group. The intrinsic differences between single- and double-stranded DNAs are related to charge, hydrophobicity, size and three-dimensional structure. These variations in biophysical properties have been utilized for comparative separations on these two resins. All deoxynucleotides and DNAs tested bound strongly to the chromatographic materials and could be eluted by a linear gradient of increasing NaCl concentration. Capto Q ImpRes provided a recognition for guanylate bases when samples of deoxynucleotides or poly(dG) were examined. This recognition was not observed for Capto Adhere. Another pronounced difference between the resins was observed in the inverted elution of ss- and dsDNA, where ssDNA eluted at 2.88 M NaCl on Capto Adhere, while on Capto Q ImpRes ssDNA eluted already at 1.47 M NaCl. This behaviour can be linked to the presence of the more hydrophobic phenyl group in Capto Adhere, leading to stronger retention of ssDNA molecules, which have a more hydrophobic character due to a higher degree of base exposure.




Similar content being viewed by others
Abbreviations
- AEX:
-
Anion exchange
- dNTP:
-
Deoxynucleotide
- dsDNA:
-
Double-stranded DNA
- HIC:
-
Hydrophobic interaction chromatography
- IgG:
-
Immunoglobulin
- MMC:
-
Multimodal chromatography
- nt:
-
Nucleotide
- pDNA:
-
Plasmid DNA
- poly(dA):
-
Polydeoxyadenylate
- poly(dC):
-
Polydeoxycytidylate
- poly(dG):
-
Polydeoxyguanylate
- poly(dT):
-
Polythymidylate
- ssDNA:
-
Single-stranded DNA
References
Sousa A, Sousa F, Queiroz JA (2012) Advances in chromatographic supports for pharmaceutical-grade plasmid DNA purification. J Sep Sci 35:3046–3058. doi:10.1002/jssc.201200307
Pereira LR, Prazeres DMF, Mateus M (2010) Hydrophobic interaction membrane chromatography for plasmid DNA purification: design and optimization. J Sep Sci 33:1175–1184. doi:10.1002/jssc.200900844
Liu MA (2011) DNA vaccines: an historical perspective and view to the future. Immunol Rev 239:62–84. doi:10.1111/j.1600-065X.2010.00980.x
Ghanem A, Healey R, Adly FG (2013) Current trends in separation of plasmid DNA vaccines: a review. Anal Chim Acta 760:1–15. doi:10.1016/j.aca.2012.11.006
Yamamoto S, Yoshimoto N, Tarmann C, Jungbauer A (2009) Binding site and elution behavior of DNA and other large biomolecules in monolithic anion-exchange chromatography. J Chromatogr A 1216:2616–2620. doi:10.1016/j.chroma.2009.01.060
Yamamoto S, Nakamura M, Tarmann C, Jungbauer A (2007) Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. J Chromatogr A 1144:155–160. doi:10.1016/j.chroma.2007.01.025
Matos T, Queiroz JA, Bülow L (2013) Binding and elution behavior of small deoxyribonucleic acid fragments on a strong anion-exchanger multimodal chromatography resin. J Chromatogr A 1302:40–44. doi:10.1016/j.chroma.2013.05.044
Ongkudon CM, Danquah MK (2010) Process optimisation for anion exchange monolithic chromatography of 4.2 kbp plasmid vaccine (pcDNA3F). J Chromatogr B 878:2719–2725. doi:10.1016/j.jchromb.2010.08.011
Sousa F, Queiroz JA (2011) Supercoiled plasmid quality assessment by analytical arginine-affinity chromatography. J Chromatogr A 1218:124–129. doi:10.1016/j.chroma.2010.11.002
Tiainen P, Galaev I, Larsson PO (2007) Plasmid adsorption to anion-exchange matrices: comments on plasmid recovery. Biotechnol J 2:726–735. doi:10.1002/biot.200700044
Mota É, Sousa Â, Černigoj U, Queiroz JA, Tomaz CT, Sousa F (2013) Rapid quantification of supercoiled plasmid deoxyribonucleic acid using a monolithic ion exchanger. J Chromatogr A 1291:114–121. doi:10.1016/j.chroma.2013.03.070
Chen WH, Fu JY, Kourentzi K, Willson RC (2011) Nucleic acid affinity of clustered-charge anion exchange adsorbents: effects of ionic strength and ligand density. J Chromatogr A 1218:258–262. doi:10.1016/j.chroma.2010.11.024
Teeters M, Root T, Lightfoot E (1036) Adsorption and desorption behavior of plasmid DNA on ion-exchange membranes: effect of salt valence and compaction agents. J Chromatogr A 2004:73–78. doi:10.1016/j.chroma.2004.03.022
Eon-Duval A, Burke G (2004) Purification of pharmaceutical-grade plasmid DNA by anion-exchange chromatography in an RNase-free process. J Chromatogr B 804:327–335. doi:10.1016/j.jchromb.2004.01.033
Diogo MM, Queiroz JA, Prazeres DMF (2003) Assessment of purity and quantification of plasmid DNA in process solutions using high-performance hydrophobic interaction chromatography. J Chromatogr A 998:109–117. doi:10.1016/S0021-9673(03)00618-6
Kallberg K, Johansson H-O, Bulow L (2012) Multimodal chromatography: an efficient tool in downstream processing of proteins. Biotechnol J 7:1485–1495. doi:10.1002/biot.201200074
Černigoj U, Vidic U, Barut M, Podgornik A, Peterka M, Štrancar A (2013) A multimodal histamine ligand for chromatographic purification of plasmid DNA. J Chromatogr A 1281:87–93. doi:10.1016/j.chroma.2013.01.058
Pezzini J, Joucla G, Gantier R, Toueille M, Lomenech A-M, Le Sénéchal C et al (2011) Antibody capture by mixed-mode chromatography: a comprehensive study from determination of optimal purification conditions to identification of contaminating host cell proteins. J Chromatogr A 1218:8197–8208. doi:10.1016/j.chroma.2011.09.036
Gilar M, Yu Y-Q, Ahn J, Fournier J, Gebler JC (2008) Mixed-mode chromatography for fractionation of peptides, phosphopeptides, and sialylated glycopeptides. J Chromatogr A 1191:162–170. doi:10.1016/j.chroma.2008.01.061
Becker K, Hallgren E, Carredano E, Palmgren R, Bülow L (2008) Characterization of multimodal hydrophobic interaction chromatography media useful for isolation of green fluorescent proteins with small structural differences. J Mol Recognit 22:104–109. doi:10.1002/jmr.897
Becker K, Grey M, Bülow L (2008) Probing protein surface accessibility of amino acid substitutions using hydrophobic interaction chromatography. J Chromatogr A 1215:152–155. doi:10.1016/j.chroma.2008.11.002
Neville DCA, Dwek RA, Butters TD (2009) Development of a single column method for the separation of lipid- and protein-derived oligosaccharides. J Proteome Res 8:681–687. doi:10.1021/pr800704t
Voitl A, Müller-Späth T, Morbidelli M (2010) Application of mixed mode resins for the purification of antibodies. J Chromatogr A 1217:5753–5760. doi:10.1016/j.chroma.2010.06.047
Kallberg K, Becker K, Bülow L (2011) Application of a pH responsive multimodal hydrophobic interaction chromatography medium for the analysis of glycosylated proteins. J Chromatogr A 1218:678–683. doi:10.1016/j.chroma.2010.11.080
Matos T, Silva G, Queiroz JA, Bulow L (2015) Preparative isolation of PCR products using mixed-mode chromatography. Anal Biochem. doi:10.1016/j.ab.2015.08.009
Montgomery DL, Prather KJ (2006) Design of plasmid DNA constructs for vaccines. Methods Mol Med 127:11–22. doi:10.1385/1-59745-168-1:11
Chakraborty S, Sharma S, Maiti PK, Krishnan Y (2009) The poly dA helix: a new structural motif for high performance DNA-based molecular switches. Nucleic Acids Res 37:2810–2817. doi:10.1093/nar/gkp133
Johnson AT, Wiest O (2007) Structure and dynamics of poly(T) single-strand DNA: implications toward CPD formation. J Phys Chem B 111:14398–14404. doi:10.1021/jp076371k
Liang X, Kuhn H, Frank-Kamenetskii MD (2006) Monitoring single-stranded DNA secondary structure formation by determining the topological state of DNA catenanes. Biophys J 90:2877–2889. doi:10.1529/biophysj.105.074104
Hebron HR, Yang Y, Hang J (2008) High performance DNA purification using a novel ion exchange matrix. J Biomol Tech 19:205–210
Westman E, Eriksson S, Laas T, Pernemalm PA, Skold SE (1987) Separation of DNA restriction fragments by ion-exchange chromatography on FPLC columns Mono P and Mono Q. Anal Biochem 166:158–171
Murphy JC, Wibbenmeyer JA, Fox GE, Willson RC (1999) Purification of plasmid DNA using selective precipitation by compaction agents. Nature 17:10–11
Acknowledgments
Tiago Matos acknowledges a fellowship (SFRH/BD/47934/2008) from FCT, Portuguese Foundation for Science and Technology. This work was supported by the Swedish Research Council (VR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Tiago Matos was funded with fellowship (SFRH/BD/47934/2008) from FCT, Portuguese Foundation for Science and Technology, and declare no conflict of interest. This study was funded by Swedish Research Council (VR). All authors declare no conflict of interest of any type.
This article does not contain any studies with animals or with human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Matos, T., Mohamed, E.T., Queiroz, J.A. et al. Capto™ Resins for Chromatography of DNA: A Minor Difference in Ligand Composition Greatly Influences the Separation of Guanidyl-Containing Fragments. Chromatographia 79, 1277–1282 (2016). https://doi.org/10.1007/s10337-016-3148-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-016-3148-3