Abstract
The Liberian Greenbul Phyllastrephus leucolepis is known only from the Cavalla Forest, Liberia, where it was seen between 1981 and 1984 but has not been found since the collection of the type, and only, specimen. It is similar to the common and widespread Icterine Greenbul P. icterinus, from which it differs primarily in having white subterminal spots on the wing coverts and all flight feathers. Its validity as a distinct species has been questioned but left unresolved. This paper describes the first genetic study of the Liberian Greenbul, to attempt to determine whether it is a distinct species, a plumage variant of Icterine Greenbul, or a hybrid. Total genomic DNA was isolated independently by two separate laboratories from the type specimen, as well as from Icterine and White-throated Greenbuls P. albigularis sampled close to the type locality in Liberia. Mitochondrial and nuclear DNA was sequenced and compared to that of Icterine and other greenbul species from Liberia and elsewhere. Sequence analysis of three mitochondrial genes and one nuclear gene showed that the Liberian Greenbul is not a hybrid but falls within the range of intraspecific genetic variation observed in the Icterine Greenbul. Reasons for this are discussed, but the most likely explanation is that the Liberian Greenbul represents a plumage variant of the Icterine Greenbul. The alternative possibilities that the Liberian Greenbul represents a distinct species which only recently diverged or has ongoing gene flow with the Icterine Greenbul cannot be formally refuted. Conservation implications for the Cavalla Forest are discussed.
Zusammenfassung
Taxonomischer Status des Fleckflügelbülbüls Phyllastrephus leucolepis und die Bedeutung des Cavalla Forest, Liberia, für seinen Schutz
Der Fleckflügelbülbül Phyllastrephus leucolepis ist nur aus dem Cavalla Forest in Liberia bekannt, wo er zwischen 1981 und 1984 beobachtet, aber seit dem Fang des Typusexemplares, des einzigen Präparates überhaupt, nicht wieder gefunden wurde. Er ist dem häufigen und weit verbreiteten Zeisigbülbül P. icterinus recht ähnlich, von dem er sich in erster Linie durch den Besitz von weißen Subterminalflecken auf den Flügeldecken und auf allen Schwingen unterscheidet. Die Anerkennung des Fleckflügelbülbüls als eigene Art wurde zwar angezweifelt, blieb aber bislang ungelöst. Diese Arbeit beinhaltet die erste genetische Untersuchung mit dem Ziel herauszufinden, ob es sich beim Fleckflügelbülbül um eine eigene Art, eine Gefiedervariante des Zeisigbülbüls oder um einen Hybriden handelt. Dazu wurden in zwei unabhängigen Labors DNA-Untersuchungen sowohl am Typusexemplar von P. leucolepis als auch an weiteren Exemplaren des Zeisigbülbüls und des Schuppenkopfbülbüls P. albigularis aus der Nähe der Typuslokalität in Liberia durchgeführt. Es wurde mitochondriale und nukleäre DNA sequenziert und mit weiteren Sequenzen des Zeisgbülbüls und anderer Bülbül-Arten aus Liberia und darüber hinaus verglichen. Die Sequenzanalyse von drei mitochondrialen und einem nukleären Gen zeigte, dass der Fleckflügelbülbül kein Hybrid ist, stattdessen aber in die intraspezifische genetische Variationsbreite des Zeisigbülbüls fällt. Mögliche Gründe werden diskutiert; die wahrscheinlichste Erklärung ist, dass der Fleckflügelbülbül eine Gefiedervariante des Zeisigbülbüls darstellt. Alternative Erklärungsansätze, nach denen der Fleckflügelbülbül eine erst kürzlich vom Zeisigbülbül abgespaltene Art ist oder die noch im Genfluss mit dem Zeisigbülbül steht, kann formal gesehen nicht ganz abgelehnt werden. Konsequenzen für den Schutz des Cavalla Forest werden diskutiert.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bulbuls and greenbuls (Aves, Passeriformes, Pycnonotidae) are medium-sized songbirds comprising approximately 130 species widely distributed across Africa and Asia. The Liberian Greenbul Phyllastrephus leucolepis is one of the world’s most poorly known bird species. It was listed as Critically Endangered until 2016, when its status was revised to Data Deficient (BirdLife International 2017a). It is known only from the Cavalla Forest, 20 km northwest of Zwedru, Grand Gedeh County in eastern Liberia, where one to two birds were seen on nine occasions by one of the authors (WG) in the dry months (November–February) of the years 1981–1984. The type specimen was collected in January 1984 and described as a species new to science (Gatter 1985). No further specimens have been obtained.
The Liberian Greenbul very closely resembles the common and widespread Icterine Greenbul Phyllastrephus icterinus in size, morphology, general plumage and tropical forest habitat. Its most conspicuous identification feature is not shown by any other Phyllastrephus species: broad white spots to all primaries, all secondaries, the larger tertials, the greater coverts, primary coverts and alula, creating two unbroken whitish wing-bars when the wing is open. All the whitish spots have dark borders right to the tips of the feathers, giving the impression of eye-spots, as shown almost perfectly on open wings in Sinclair and Ryan (2003) (Fig. 1a). Additionally, in comparison to typical Icterine Greenbuls, the tail of the Liberian Greenbul specimen is slightly paler, the wing is somewhat redder-brown and less olive, and there are very dark brown rather than black inner webs to the flight feathers (Gatter 1985).
External morphology of the Liberian Greenbul. a Artistic representations of the Liberian Greenbul Phyllastrephus leucolepis in life. The most obvious feature that distinguishes it from the otherwise very similar Icterine Greenbul P. icterinus is the presence of white subterminal ‘eye spots’ to the wing feathers—a feature not shown by any other species of Phyllastrephus. Upper painting reproduced from Gatter (1985); lower painting by Norman Arlott reproduced from Sinclair and Ryan (2003) with permission. b The type specimen of Liberian Greenbul. Its poor condition is due to damage by ants, resulting from the interval of a night between it being shot and its subsequent retrieval from foliage
The species has not been seen since the type was collected in 1984. The civil wars that Liberia suffered between 1989 and 2003 rendered expeditions to this area impossible. Even after 2003, Ivorian rebels retreated to the border area as a place of refuge. Because of UN attacks on rebel-held coastal towns that were used to export logs, the rebels moved their logging activities further inland to the northeastern border area, which gave them access to ports in neighbouring Ivory Coast. This had serious consequences for the forests in question.
Targeted searches of the two known sites for five days during the rainy season in July 2010 were unsuccessful (Molubah and Garbo 2010). An expedition from 7 February to 8 March 2013, taking in the type locality, also failed to locate the species (Phalan et al. 2013). The Cavalla Forest is of global conservation significance. Following the publications of Gatter (1988, 1992, 1997), Gatter and Gardner (1993) and Gatter et al. (1988), it was recognized as an Important Bird and Biodiversity Area by BirdLife International, not only for Liberian Greenbul but also for the presence of other globally threatened bird species, including the Vulnerable White-breasted Guineafowl Agelastes meleagrides and Brown-cheeked Hornbill Bycanistes cylindricus (Robertson 2001). Globally threatened mammals also occur, including Chimpanzee Pan troglodytes (Endangered), while Western Red Colobus Procolobus badius and Pygmy Hippopotamus Choeropsis liberiensis, both also Endangered, are thought to persist (Phalan et al. 2013). Biodiversity in this area is, however, threatened.
Commercial logging is not allowed at present, but larger bodied species are subject to sustained hunting pressure. Despite being tentatively accepted as a valid species by many authors, the status of Liberian Greenbul has been considered unclear, requiring further investigation (e.g., Vuilleumier et al. 1992; Fishpool and Tobias 2005; del Hoyo and Collar 2016). Here, we consider three hypotheses: first, that it is genuinely a distinct species of Phyllastrephus; second, that it is an aberrant plumage variation of Icterine Greenbul, caused either by genetic mutation or environmental factors such as nutritional deficiency; and third, that it is a hybrid. The only sympatric greenbul with pale spots on its wings is the Spotted Greenbul Ixonotus guttatus.
In the absence of any further field observations of the Liberian Greenbul, a genetic study had the potential to resolve its taxonomic status. Tissue samples were hence obtained separately and sent for independent sequencing by JMC and MP. This paper describes the results of the genetic analyses that lead us to the conclusion that the Liberian Greenbul is most likely a plumage variant of the Icterine Greenbul.
Methods
Sampling
The Liberian Greenbul Expedition, led by BP and LF, visited the Cavalla Forest between 7 February and 8 March 2013, searching and mist-netting at several locations within a search area of approximately 10 × 15 km, 20 km northwest of Zwedru, as far as the border with Ivory Coast. Surveys were focused in the vicinity of the type locality of Liberian Greenbul. No Liberian Greenbuls were observed, but Icterine Greenbuls were common, and blood samples of less than 200 μl were taken from the brachial veins of 17 birds using a fine glass capillary, and stored in ethanol. Sampling was undertaken with permission from the Forestry Development Authority, and all sampled birds were kept for 5 min before release to ensure full recovery with no bleeding. A single White-throated Greenbul P. albigularis was also sampled for DNA analysis.
The type specimen of Liberian Greenbul is at the Museum Alexander Koenig, Bonn (accession number ZFMK 84.221; van den Elzen 2010) (Fig. 1b). Toepads were isolated by museum staff and sent to JMC and MP for independent DNA isolation and analysis.
DNA isolation
At the Aberdeen lab (JMC), total genomic DNA was isolated from toepads and blood samples using the QIAGEN DNA Micro Kit (QIAGEN, UK) according to the manufacturer’s instructions with the addition of dithiothreitol to a concentration of 0.1 M in the overnight proteinase K digestion. Elution was in 80 μl buffer AE. Isolation of Liberian Greenbul DNA and initial PCRs were performed before any other Phyllastrephus material was received or handled, thus there was no possibility of contamination. At the Dresden lab (MP), total genomic DNA was isolated from toepads using the sbeadex® forensic kit (LGC Genomics) according to the manufacturer’s instructions except for overnight (instead of 1 h) incubation of tissue with proteinase K and an elution volume of only 60 µl (instead of 100 µl). To avoid cross-contamination with fresh DNA, extraction and PCR were performed in a separate clean lab on separate working benches at each step in the analysis (sampling, extraction and PCR). After each step, the working benches were cleaned with DNA-away (Molecular Bio Products, Inc.) and both benches and lab rooms were decontaminated with UV light for at least 4 h.
PCR and sequencing
To isolate full-length fragments for genes encoding cytochrome b (cytb), cytochrome oxidase subunit 1 (COI) and NADH:ubiquinone oxidoreductase core subunit 2 (ND2) from DNA from blood samples, the universal bird PCR primers L14993/H16065, BirdF1/BirdR1 and L5216/H6313, respectively (Helbig et al. 1995; Hebert et al. 2004; Shannon et al. 2014), were employed using Bio-X-Act Short DNA polymerase (Bioline, UK) with PCR reactions and conditions as described previously (Helbig et al. 1995; Hebert et al. 2004; Shannon et al. 2014; Dejtaradol et al. 2016 ). Although the Liberian Greenbul specimen was only 33 years old, none of the above primers produced bands from that sample, consistent with significant DNA degradation. Primers were designed to amplify short fragments (120–200 bp) of these genes from degraded DNA by aligning available sequences from multiple Phyllastrephus species (downloaded using NCBI Gene—https://www.ncbi.nlm.nih.gov/gene) using CLC Sequence Viewer (QIAGEN Bioinformatics) and choosing primers in highly conserved regions. These primers are detailed in Table 1.
In Aberdeen (JMC), PCR reactions were run on 1.5% agarose gels in TAE buffer and bands of the predicted size were cut out. DNA was purified using the QIAGEN gel extraction kit (QIAGEN, UK) according to the manufacturer’s instructions, with resuspension in 30 μl of buffer EB. Sequencing was performed by Source Bioscience (Nottingham, UK). In Dresden (MP), PCR products were purified using ExoSAP-IT (GE Healthcare) according to the manufacturer’s instructions and sequenced on an ABI 3130xl capillary sequencer (Applied Biosystems).
All PCR products were sequenced on both strands, with any inconsistencies resolved by manual inspection of chromatograms or, if necessary, resequencing. Verified sequences were uploaded to the European Nucleotide Archive with accession numbers LT898200-LT898220 and MF362667.
DNA analysis and phylogenetic trees
DNA sequences were aligned using CLC Sequence Viewer v6 (http://www.clcbio.com/products/clcsequence-viewer/), inspected by eye, and exported as fasta files. Maximum likelihood phylogenetic trees were generated from alignments using PhyML (Dereeper et al. 2008) online via the South of France Bioinfomatics Platform (http://www.atgc-montpellier.fr/index.php?type=pg). The default substitution model was used, which for PhyML is HKY85 (Hasegawa et al. 1985; Guindon et al. 2010) with a transition/transversion ratio of 4.0, a gamma shape parameter of 1.0, and assuming no invariable sites. A BIONJ distance-based starting tree was refined by PhyML using the maximum likelihood algorithm. Trees were rendered by TreeDyn (Chevenet et al. 2006). Taxon sampling is heterogeneous across different trees because few greenbul species have been sampled at all loci.
Results
A 358-bp fragment of the ‘barcode’ marker, COI, was obtained from the Liberian Greenbul sample independently by both groups and compared to sequences obtained from Icterine Greenbuls from Liberia, downloaded Genbank sequences of P. icterinus from elsewhere in Africa, and sequences from White-throated Greenbul P. albigularis (also sampled in Liberia). The Liberian Greenbul sequence was identical to that from one of the Icterine Greenbuls (‘MAR6611’) sampled in Liberia. As a whole, the P. icterinus and P. leucolepis sequences formed a clade with only 3/358 variable nucleotides (0.8% uncorrected divergence), in contrast to the 21 sites separating the icterinus/leucolepis group from P. albigularis (5.8% uncorrected divergence), a species identified as being closely related to Icterine Greenbul in previous molecular analyses (Moyle and Marks 2006; Johansson et al. 2007; Shakya and Sheldon 2017). These data suggested that the Liberian Greenbul falls within the range of genetic variation of Icterine Greenbul from Liberia and elsewhere (Fig. S1 in the Electronic supplementary material, ESM).
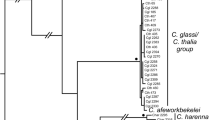
In contrast to COI, the cytb gene has been widely sequenced in greenbuls. Full length cytb sequences were therefore obtained from Icterine and White-throated Greenbuls sampled in Liberia, and multiple database cytb sequences from these and other Phyllastrephus species. These were compared to 304 bp of the cytb sequence obtained from the leucolepis sample using bespoke primers designed to amplify a short fragment of mtDNA (Table 1). The Liberian Greenbul sequence was 1 bp different from an Icterine Greenbul from Liberia sequenced by us (‘Ict06’) and from a database sequence of Icterine Greenbul from Congo (AF199382). For cytb as a whole, there were 3 variable sites within the icterinus/leucolepis clade (1% uncorrected divergence) compared with 26 bp separating them from albigularis. The leucolepis/icterinus clade had 99/100 bootstrap support to the exclusion of all other taxa and there was no evidence of any genetic distinctiveness of leucolepis (Fig. 2a).
Genetic status of Liberian Greenbul. a Gene tree (maximum likelihood) based on 308 bp of cytochrome b (cytb) for multiple greenbul taxa, including the Liberian, Icterine and White-throated Greenbuls sequenced in this study. Bootstrap support (100 replicates) for key nodes is indicated. The topology of much of the tree is poorly resolved because of the short stretch of sequence yielded by degraded genomic DNA in the Liberian specimen. However, the clade containing closely related Icterine and Liberian Greenbuls, as a sister to albigularis and to the exclusion of all other taxa, is strongly supported. Nomenclature follows Dickinson and Christidis (2014). Accession numbers for species downloaded from NCBI (https://www.ncbi.nlm.nih.gov/gene) are given. b Gene tree (maximum likelihood) based on NADH:ubiquinone oxidoreductase core subunit 2 (ND2) for multiple greenbul taxa, including the Liberian, Icterine and White-throated Greenbuls sequenced in this study. Bootstrap support (100 replicates) for key nodes is indicated. Yellow shaded area represents the presumed nuclear allele of ND2 sequenced incidentally during this study. P. Phyllastrephus
Analysis of the ND2 sequence was complicated by the presence of a presumed nuclear copy of the gene (nuND2). ‘Universal’ primers (described in “Methods”) yielded full-length ND2 from multiple samples of Icterine Greenbul and White-throated Greenbul, but gave no product from Liberian Greenbul DNA. The ND2 sequences obtained from Icterine and White-throated Greenbuls were near-identical to database sequences obtained by other groups for these species and gave predicted translations yielding full-length ND2 homologous to the mitochondrial protein in other bird taxa. However, some bespoke primers designed to amplify short fragments of ND2 from Phyllastrephus species (Table 1) yielded sequences from Liberian, Icterine and White-throated Greenbuls that were divergent from the mitochondrial gene but with much less intraspecific divergence than the mitochondrial allele. We postulate that this is a nuclear copy and tentatively label it nuND2. 599 bp of the presumed nuND2 were isolated from Liberian Greenbul, in two overlapping fragments isolated separately by the two groups. These data are represented in Fig. S2 in the ESM. The presumed nuND2 appears to be highly conserved: the two icterinus sequences isolated were identical, 1 bp different from the leucolepis allele, with a further 2 bp distance to albigularis. Bespoke primers designed subsequently to amplify the mitochondrial ND2 gene from Phyllastrephus, but which had mismatches compared to the sequence from the presumed nuclear allele (YMLibF2 and YMLibR3—Table 1), amplified a 199-bp fragment from Liberian and Icterine Greenbuls that was identical to the same 199-bp subset of the full-length mitochondrial sequence of Icterine Greenbul isolated using universal primers L5216 and H6313. We therefore believe this fragment to represent part of the true mitochondrial ND2 of Liberian Greenbul. A gene tree including this fragment is presented in Fig. 2b. Based on a short fragment of sequence, many of the nodes in the tree have equivocal bootstrap support, and it does not represent a robust greenbul phylogeny. However, the Liberian Greenbul allele was identical to one of the alleles of Icterine Greenbul sampled in Liberia, within 3 bp of all other Icterine Greenbuls sampled in Liberia, and showed 193/199-bp identity with the Icterine Greenbul sampled in Cameroon. At least 17 bp (>11% uncorrected sequence divergence) separated Icterine/Liberian Greenbuls from all other sampled species. The data are consistent with the hypothesis that the Liberian Greenbul falls within the range of intraspecific variation in the Icterine Greenbul. A concatenated tree based on 846 bp of COI, cytb and ND2 sequences from Icterine, Liberian and White-throated Greenbuls (the only relevant taxa for which all genes are available) was consistent with this conclusion (Fig. S3 in the ESM).
Mitochondrial sequences give no information about the male parents of sampled birds, and phylogenies based on mitochondrial data alone may not accurately reconstruct the evolutionary history of taxa. To determine whether nuclear DNA also suggested a relationship with Icterine Greenbul and whether Liberian Greenbul represents a hybrid between Icterine and Spotted Greenbuls, 176 bp was amplified and sequenced from intron 5 of the β-fibrinogen gene of the Liberian Greenbul sample, as well as from an Icterine Greenbul collected in Liberia in 2013 (ICT05), and aligned to database sequences of the same intron from Spotted Greenbul and multiple other Phyllastrephus species. The Liberian Greenbul sequence was identical to that of the Icterine Greenbul from Liberia with the exception of one position (see Fig. S4 of the ESM), which was heterozygous C/T in the Liberian Greenbul but homozygous C/C in Icterine Greenbul ‘ICT05’. This single SNP difference was verified in forward and reverse sequences. Both the Icterine Greenbul ‘ICT05’ and the Liberian Greenbul additionally differed only at position 8 (A/A) from the downloaded Icterine Greenbul sample with accession number EF626731 (A/A).
In contrast, there were 9 single-bp changes and a 2-bp indel between Icterine/Liberian Greenbul sequences and that of Spotted Greenbul. That the Liberian Greenbul was heterozyogous at none of these positions strongly suggests that it was not an Icterine × Spotted Greenbul hybrid.
Discussion
The current study has failed to find any significant genetic distance between Icterine and Liberian Greenbuls, whilst demonstrating large and diagnostic genetic distances between other species of greenbul in the genus Phyllastrephus. Previous molecular studies of bulbuls, including greenbuls, showed typically high levels of genetic divergence between different species (5–10% uncorrected at mitochondrial loci) (Moyle and Marks 2006; Shakya and Sheldon 2017). The data are limited by the fact that only one specimen of Liberian Greenbul has ever been obtained, and the fact that sampling of other greenbul species is very incomplete—much more work would be required to produce a robust, complete greenbul phylogeny. However, the clearest result from the data reported here is that the Liberian Greenbul falls within the range of genetic variation observed among Icterine Greenbuls in Liberia and elsewhere. Although a hybrid hypothesis seemed inherently highly improbable, and intergeneric hybridisation is unknown in greenbuls, we have also excluded the possibility that the Liberian Greenbul is a hybrid between Icterine and Spotted Greenbuls.
Having excluded a hybrid hypothesis, the lack of any observed significant genetic difference between Icterine Greenbul and the Liberian Greenbul specimen suggests one of three theoretical possibilities:
-
(1)
Icterine and Liberian Greenbuls are conspecific and the Liberian Greenbul is or was a local plumage variant of Icterine Greenbul.
-
(2)
Icterine and Liberian Greenbuls are separate or incipient species but have diverged very recently, such that ancestral gene alleles are shared.
-
(3)
The Icterine and Liberian Greenbuls are separate species with a long period of divergence but with a degree of gene flow such that the leucolepis specimen carried introgressed Icterine gene alleles.
Our data point strongly to the likelihood that Liberian Greenbul represents a plumage variety of Icterine Greenbul. This conclusion is analogous to the case of the Bulo Burti Boubou Laniarius liberatus, described as new to science on the basis of a single individual from Somalia (Smith et al. 1991). It was later shown to fall within the range of genetic variation in Somali Boubou L. erlangeri and is now regarded as a rare colour morph of that species (Nguembock et al. 2008).
Of the alternative hypotheses, the introgression of both mitochondrial and nuclear DNA suggested by the current study make the third possibility above extremely implausible. The second possibility (that the Liberian Greenbul is a valid taxon that has only recently diverged) also now seems unlikely. The Liberian Greenbul is, or was, sympatric with Icterine Greenbul and, indeed, Icterine Greenbuls were sampled by us recently at the site where the Liberian Greenbul specimen was obtained (Phalan et al. 2013). Aside from Icterine, Liberian and White-throated Greenbuls, there are no other species of Phyllastrephus in the Cavalla Forest and Zwedru area, though an additional 11 greenbul species do occur in the genera Andropadus, Calyptocichla, Baeopogon, Chlorocichla, Thescelocichla and Ixonotus (the Spotted Greenbul I. guttatus described above). A significant degree of niche separation would be expected between two or more separate species cohabiting stably in the same habitat. Such syntopy without evidence of niche separation, and with gene flow and/or retained ancestral alleles, would be unstable and very unusual. Ecological segregation is usually accompanied by appreciable morphological, vocal, genetic and/or behavioural differentiation. If two closely related sympatric taxa were to maintain such differences over long periods of time, this would be taken as extremely strong evidence for separate species status (Helbig et al. 2002). While nothing is known of the voice of the Liberian Greenbul, in terms of general morphology, biometrics and plumage coloration it is almost identical to the Icterine Greenbul, except for the distinctive white spots to the greater coverts and remiges of P. leucolepis, which are absent in P. icterinus. There are several possible explanations for the white spots. Were the Liberian Greenbul a valid species, they could potentially have a role in signalling species recognition, facilitating a pre-mating reproductive barrier between the two species or incipient species. In support of this, WG observed Liberian Greenbuls raising and quivering their wings, displaying their white spots in a manner suggestive of intraspecific signalling, and in a manner different from that observed in Icterine Greenbuls, which also habitually extend and raise one or both wings. This behaviour, together with the plumage differences and multiple observations in different years and at two locations 2 km apart, was a deciding factor in recognising Liberian Greenbul as a species (Gatter 1985). However, it is also possible that the white spots represent a plumage variation of Icterine Greenbul. This possibility, as discussed by Gatter (1985), could occur because of a genetic mutation or because of nutritional deficiency during feather growth, leading to a loss of pigmentation at the tips of the wing feathers. For the latter explanation to hold true, the consistency of patterning within and across feather tracts suggests that all of the feathers grew simultaneously, as would be expected in juvenile, but not in adult, plumage.
Formal resolution of the status of the Liberian Greenbul will not be possible until either a population of Liberian Greenbuls is rediscovered or alternatively shown not to exist. Such a population is apparently no longer extant at the type locality. We have considered whether Icterine or Liberian Greenbuls may be nomadic or migratory. All observations of Liberian Greenbuls were restricted to the timespan between the last days of October and February, which may support the hypothesis that they represent aberrant juveniles. The expedition of February 2013 mistnetted a juvenile Icterine Greenbul as well as a female with a brood patch, consistent with the dry season observations of Liberian Greenbul. Many rainforest species in the region, including greenbuls, show a marked seasonality in breeding; they are in breeding condition from the second half of the rainy season and have independent young from the end of it (Mattes and Gatter 1989). However, this alone would not explain why the observations of leucolepis are restricted to a time period of three months. Data on local movements or migration of the lowland forest greenbuls of the region are lacking. Although there is no evidence of significant seasonal migrations of Icterine or any other greenbul species into or out of the study area, the distributional area of the greenbuls in Liberia extends over the region of the Guinea wet savannah as well as that of the rainforest south of it (Borrow and Demey 2001). Any migration is therefore difficult to confirm. One bulbul species, Yellow-throated Leaflove Chlorocichla flavicollis, is restricted to the vegetation belt of the Guinea wet savannah and does not breed in the forest zone. However, since 2005 there have been dry-season (January/February) records from five locations in Liberian open coastal habitats 200–300 km south of its known range (M. Fischer, F. Molubah, WG), suggesting that at least some bulbul species do undertake some dry season movements.
Liberia has experienced very significant deforestation, impacting on bird species composition (Kofron and Chapman 1995). The Liberian Greenbul expedition found that extensive areas of apparently suitable forest habitat remain in the vicinity of the type locality. Some species associated with high forest, such as Shining Drongo Dicrurus atripennis, appear to have declined since the 1980s (Phalan et al. 2013). A comparison between the species lists of Phalan et al. (2013) and Gatter (1997) shows the substantial change in species composition from a primary forest bird community to one of secondary forest with elements of farmbush and treefall gaps.
The area, including two of eight former research plots from the 1980s, was revisited by WG in July 2016 along a 70-km strip of forest bordering Ivory Coast to either side of the only border crossing to Ivory Coast. Compared with the 1980s, there is now a dense network of logging roads dating from the civil war. The once common Gola Malimbe Malimbus ballmanni (the only large population ever found) was not seen at all.
In spite of ongoing threats from—in particular—commercial logging, but also hunting, agricultural incursion (a large area of clearcut was found in 2016 at one of the former research plots where leucolepis had been found), and artisanal mining operations, the Cavalla Forest continues to support at least 20 birds and mammals on the global Red List of Threatened Species. That there are still apparently healthy populations of White-breasted Guineafowl Agelastes meleagrides (Vulnerable) and other Upper Guinea endemics is a demonstration of the importance of the area for conservation (Phalan et al. 2013). Even if the Liberian Greenbul is considered a local plumage variety of Icterine Greenbul (classified as Least Concern by BirdLife International 2017b), the conservation significance of the Cavalla Forest remains extremely high.
References
BirdLife International (2017a) Species factsheet: Phyllastrephus leucolepis. http://datazone.birdlife.org/species/factsheet/22712975. Accessed 5 Mar 2017
BirdLife International (2017b) Species factsheet: Phyllastrephus icterinus. http://datazone.birdlife.org/species/factsheet/Icterine-Greenbul. Accessed 5 Mar 2017
Borrow N, Demey R (2001) Birds of western Africa. Christopher Helm, London
Chevenet F, Brun C, Banuls AL, Jacq B, Chisten R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform 7:439
Dejtaradol A, Renner SC, Karapan S, Bates PJJ, Moyle RG, Päckert M (2016) Indochinese–Sundaic faunal transition and phylogeographical divides north of the Isthmus of Kra in Southeast Asian Bulbuls (Aves: Pycnonotidae). J Biogeogr 43:471–483
Del Hoyo J, Collar NJ (2016) HBW and BirdLife International illustrated checklist of the birds of the world, Passerines, vol 2. Lynx Edicions, Barcelona
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36(Web Server issue): W465–W469
Dickinson EC, Christidis L (eds) (2014) The Howard and Moore complete checklist of the birds of the world, vol 2, 4th edn. Aves, Eastbourne
Fishpool LDC, Tobias JA (2005) Liberian Greenbul Phyllastrephus leucolepis. In: Del Hoyo J, Elliot A, Christie D (eds) Handbook of the birds of the world, vol. 10: Cuckoo-Shrikes to Thrushes. Lynx Edicions, Barcelona, p 215
Gatter W (1985) Ein neuer Bülbül aus Westafrika (Aves, Pycnonotidae). J Ornithol 126:155–161
Gatter W (1988) The birds of Liberia. A preliminary check-list with status and open questions. Verh orn Ges Bayern 24:689–723
Gatter W (1992) White Breasted Guineafowl. World Pheas Assoc News 36:27–28
Gatter W (1997) Birds of Liberia. Pica, Mountfield
Gatter W, Gardner R (1993) The biology of the Gola Malimbe Malimbus ballmanni Wolters 1974. Bird Conserv Int 3:87–103
Gatter W, Peal A, Steiner C, Weick F (1988) The unknown immature plumages of the rare White-breasted Guinea Fowl (Agelastes meleagrides Bonaparte, 1850). Ökol d Vögel 10:105–111
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial-DNA. J Mol Evol 22:160–174
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004) Identification of birds through DNA barcodes. PLoS Biol 2:e312
Helbig AJ, Seibold I, Martens J, Wink M (1995) Genetic differentiation and phylogenetic relationships of Bonelli’s Warbler Phylloscopus bonelli and Green Warbler P. nitidus. J Avian Biol 26:139–153
Helbig AJ, Knox AG, Parkin DT, Sangster G, Collinson M (2002) Guidelines for assigning species rank. Ibis 144:518–525
Johansson US, Fjeldså J, Lokugalappatti LGS, Bowie RCK (2007) A nuclear DNA phylogeny and proposed taxonomic revision of African greenbuls (Aves, Passeriformes, Pycnonotidae). Zoolog Scr 36:417–427
Kofron CP, Chapman A (1995) Deforestation and bird species composition in Liberia, West Africa. Trop Zool 8:239–256
Mattes H, Gatter W (1989) Jahresperiodik und Biometrie von Andropadus latirostris in Liberia. Annual cycle and biometrics of Andropadus latirostris in Liberia. Stuttgarter Beitr Naturk Ser A 429:1–10
Molubah FP, Garbo M (2010) Liberian Greenbul—Phyllastrephus leucolepis. Survey report. Society for the Conservation of Nature of Liberia, Monrovia
Moyle RG, Marks BD (2006) Phylogenetic relationships of the bulbuls (Aves: Pycnonotidae) based on mitochondrial and nuclear DNA sequence data. Mol Phylogenet Evol 40:687–695
Nguembock B, Fjeldså J, Couloux A, Pasquet E (2008) Phylogeny of Laniarius: molecular data reveal L. liberatus synonymous with L. erlangeri and “plumage coloration” as unreliable morphological characters for defining species and species groups. Mol Phylogenet Evol 48:396–407
Phalan B, Fishpool LDC, Loqueh EM, Grimes T, Molubah FP, Garbo M (2013) Liberian Greenbul Expedition 2013: final report. Report to African Bird Club and RSPB. https://www.africanbirdclub.org/sites/default/files/2012_Liberian_Greenbul.pdf
Robertson P (2001) Liberia. In: Fishpool LDC, Evans MI (eds) Important bird areas in Africa and associated islands: priority sites for conservation. Pisces/BirdLife International, Newbury/Cambridge, pp 473–480
Shakya SB, Sheldon FH (2017) The phylogeny of the world’s bulbuls (Pycnonotidae) inferred using a supermatrix approach. Ibis 159:498–509
Shannon TJ, McGowan RY, Zonfrillo B, Piertney S, Collinson JM (2014) A genetic screen of the island races of Wren Troglodytes troglodytes in the North-east Atlantic. Bird Study 61:135–142
Sinclair I, Ryan P (2003) A comprehensive illustrated field guide: birds of Africa south of the Sahara. Struick, Cape Town
Smith EFG, Arctander P, Fjeldså J, Amir O (1991) A new species of shrike (Laniidae: Laniarius) from Somalia, verified by DNA sequence data from the only known individual. Ibis 133:227–235
van den Elzen R (2010) Type specimens in the bird collections of the Zoologisches Forschungsmuseum Alexander Koenig, Bonn. Bonn Zool Bull 59:29–77
Vuilleumier F, LeCroy M, Mayr E (1992) New species of birds described from 1981 to 1990. Bull Br Orn Club Suppl 112A:267–309
Acknowledgements
We thank Jochen Martens for his long-lasting patience in dealing with the specimen of leucolepis, and Brian Hillcoat for comments and advice. It is hardly possible to thank by name all those who have supported WG over the past 30 years and more since 1981 in the fields of forest ecology and ornithology in eastern Liberia. In particular, we express gratitude to Alex Peal and Theo Freeman, both Heads of Wildlife and National Parks, for their many years of cooperation, and the Silviculture Officers Wynn Bryant, Momo Kromah and Steve Miapeh. The knowledge of the tree experts Joe Keper and Daniel Dorbor helped us to gain insights into the ecological complexities of the relationship between man, birds and trees. William Toe worked for three years as bird trapper and assistant in bird banding. WG’s attachment to the University of Liberia and to the students who so often accompanied him was made possible by Ben Karmorh from the Environmental Protection Agency (EPA) and University of Liberia. NABU, the German Conservation Society, has supported the Liberian projects for almost 30 years now. We also thank Nigel Collar, Françoise Dowsett-Lemaire and Hannah Rowland for comments and advice. We thank the African Bird Club and the Royal Society for the Protection of Birds for helping to fund the 2013 expedition to the Cavalla Forest, in particular Alice Ward-Francis, Robert Sheldon, Alan Williams and Keith Betton. We also are extremely grateful to Michael Garbo and staff of the Society for the Conservation of Nature in Liberia for all manner of help with the expedition, to Harrison Karnwea and colleagues at the Forest Development Authority of Liberia for permissions and other support, as well as to Emmanuel Loqueh, Trokon Grimes, Flomo Molubah and Amos ‘Dweh’ Dorbor for being such excellent companions in the field. YL performed the genetic work as part of her M.Sc. (Genetics) at the University of Aberdeen, whose support is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare they have no competing interests. Blood sampling was undertaken with permission from the Liberian Forestry Development Authority. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Communicated by J. T. Lifjeld.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Collinson, J.M., Päckert, M., Lawrie, Y. et al. Taxonomic status of the Liberian Greenbul Phyllastrephus leucolepis and the conservation importance of the Cavalla Forest, Liberia. J Ornithol 159, 19–27 (2018). https://doi.org/10.1007/s10336-017-1477-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-017-1477-0